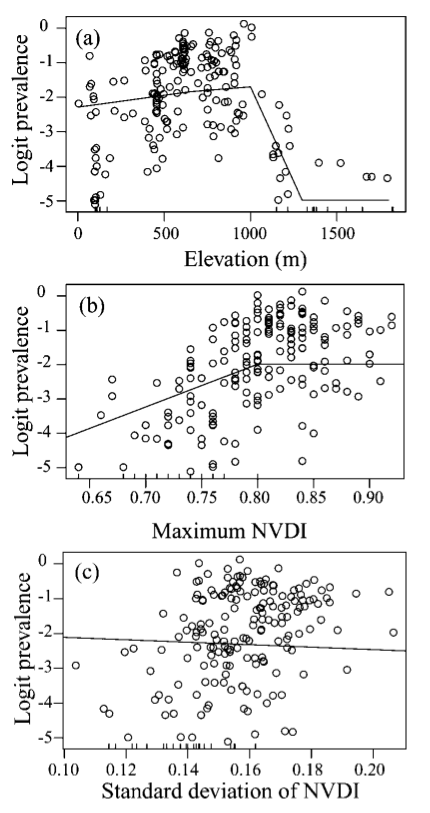
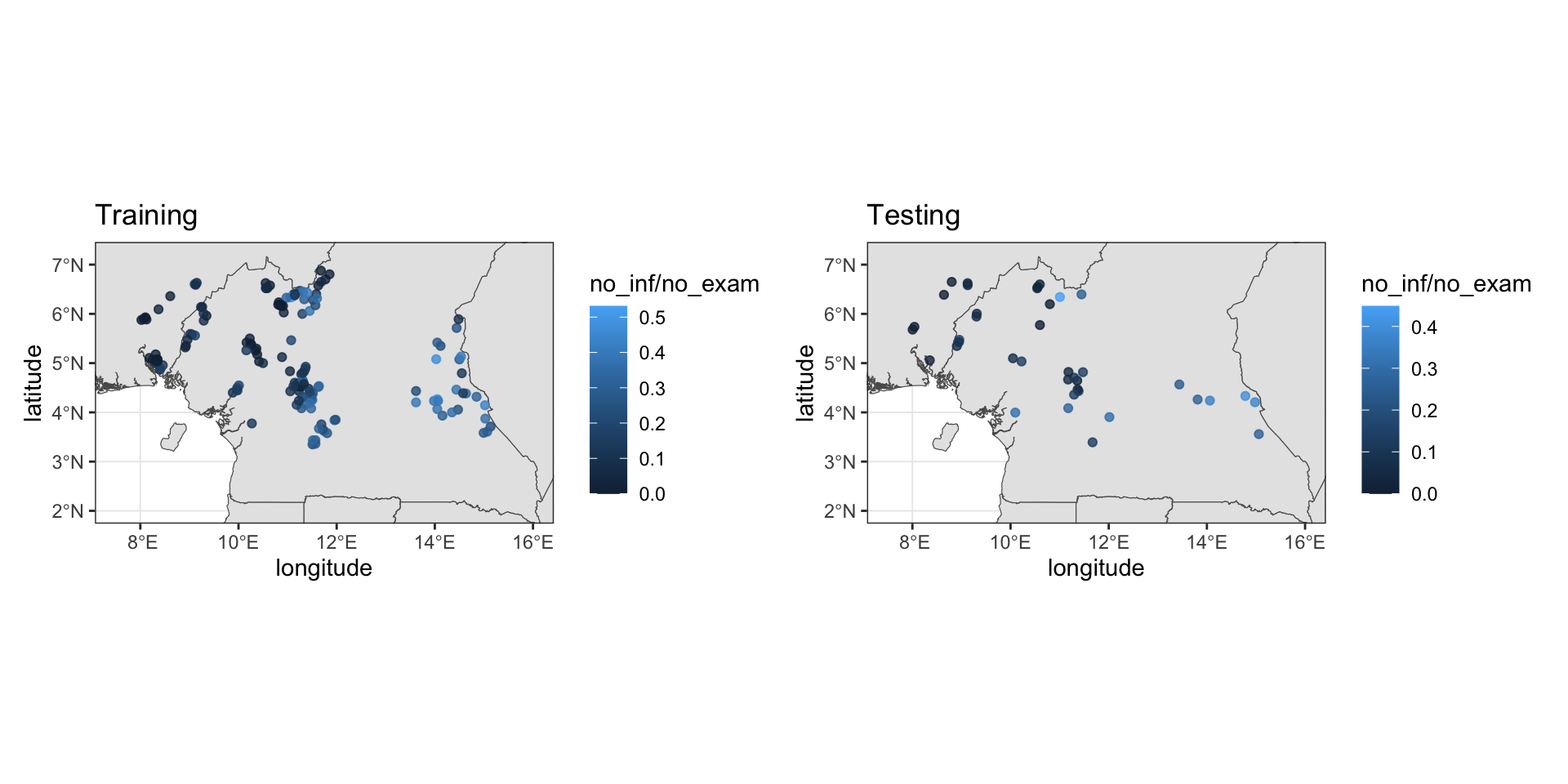
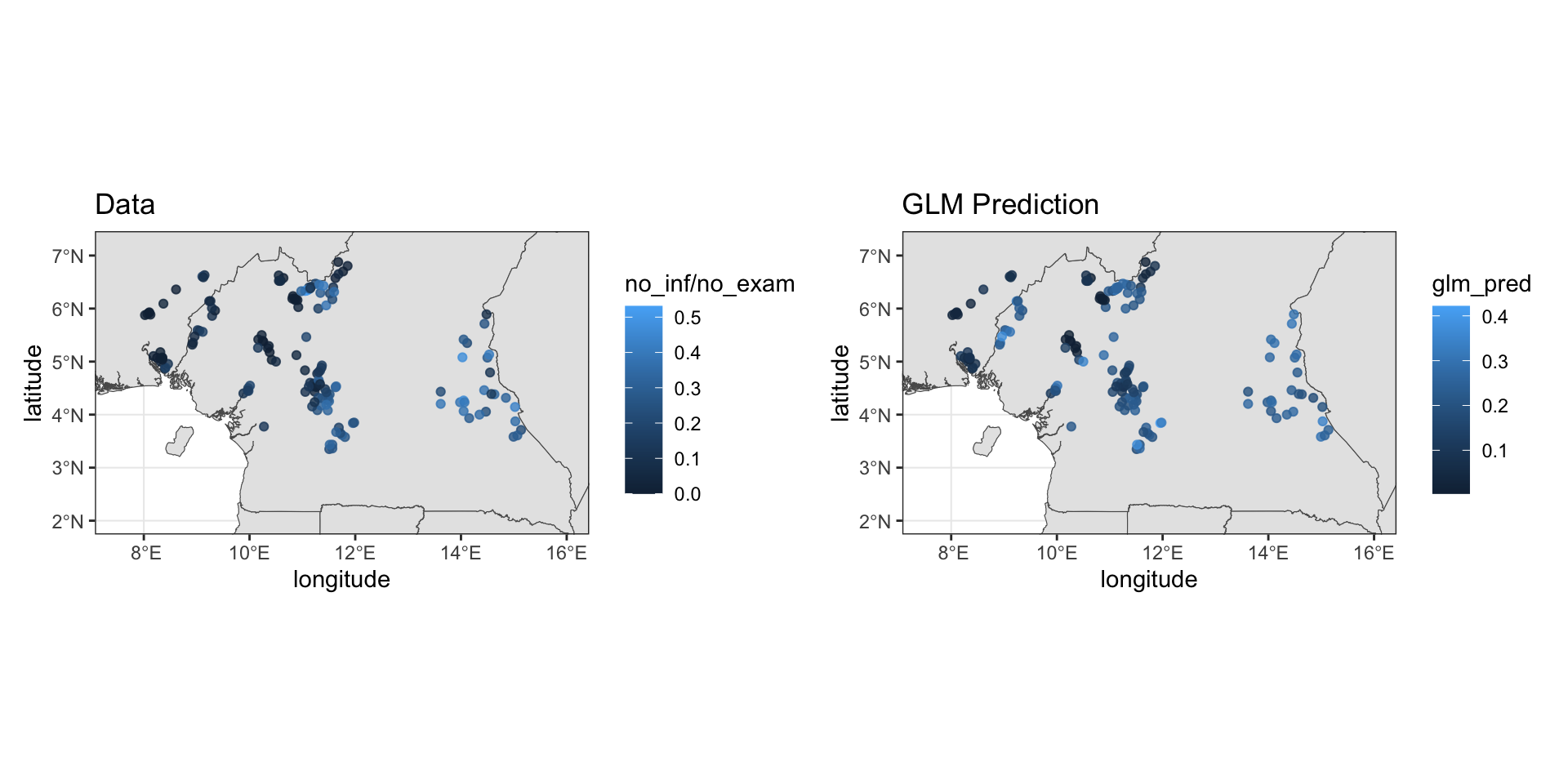
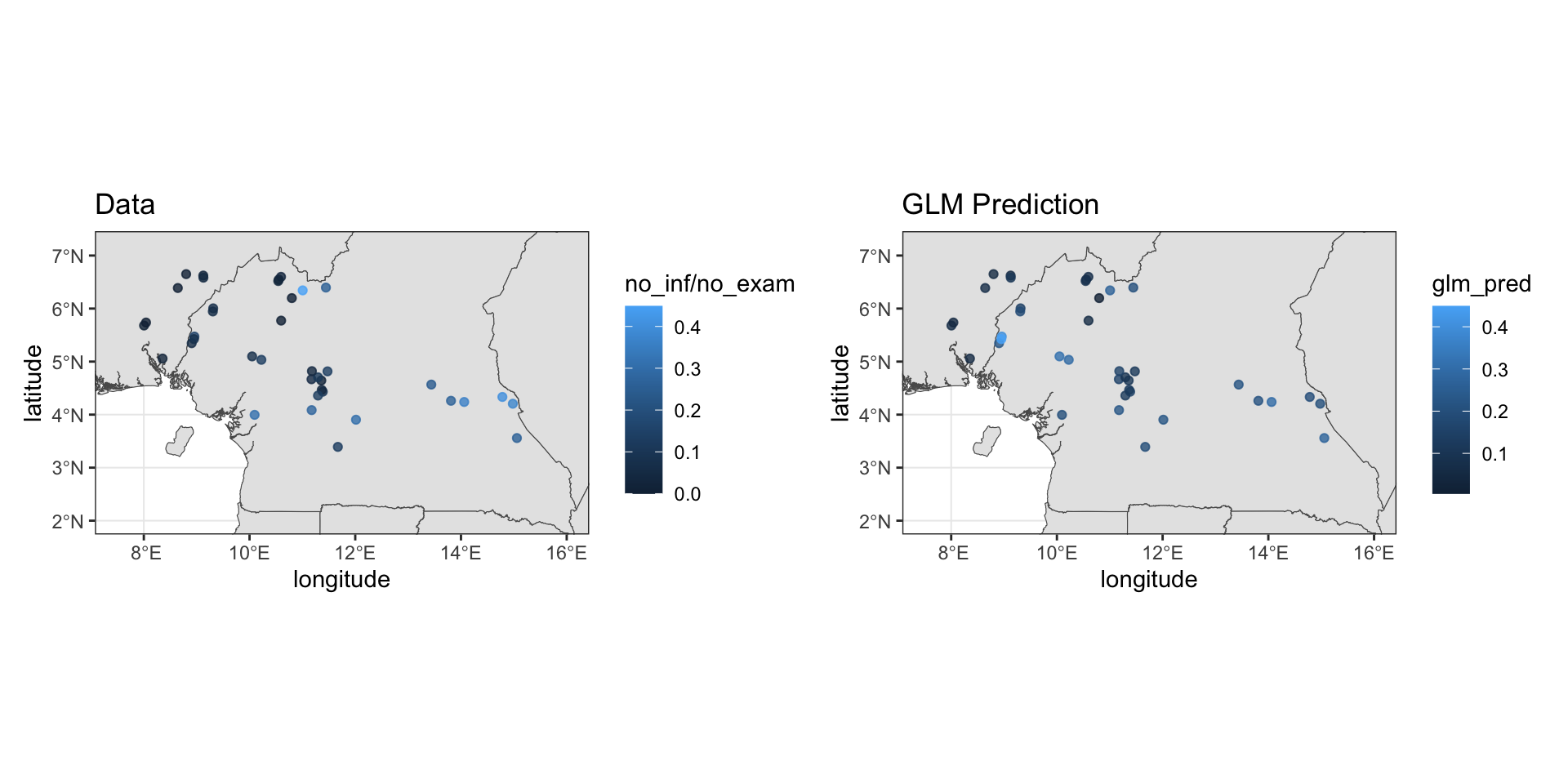
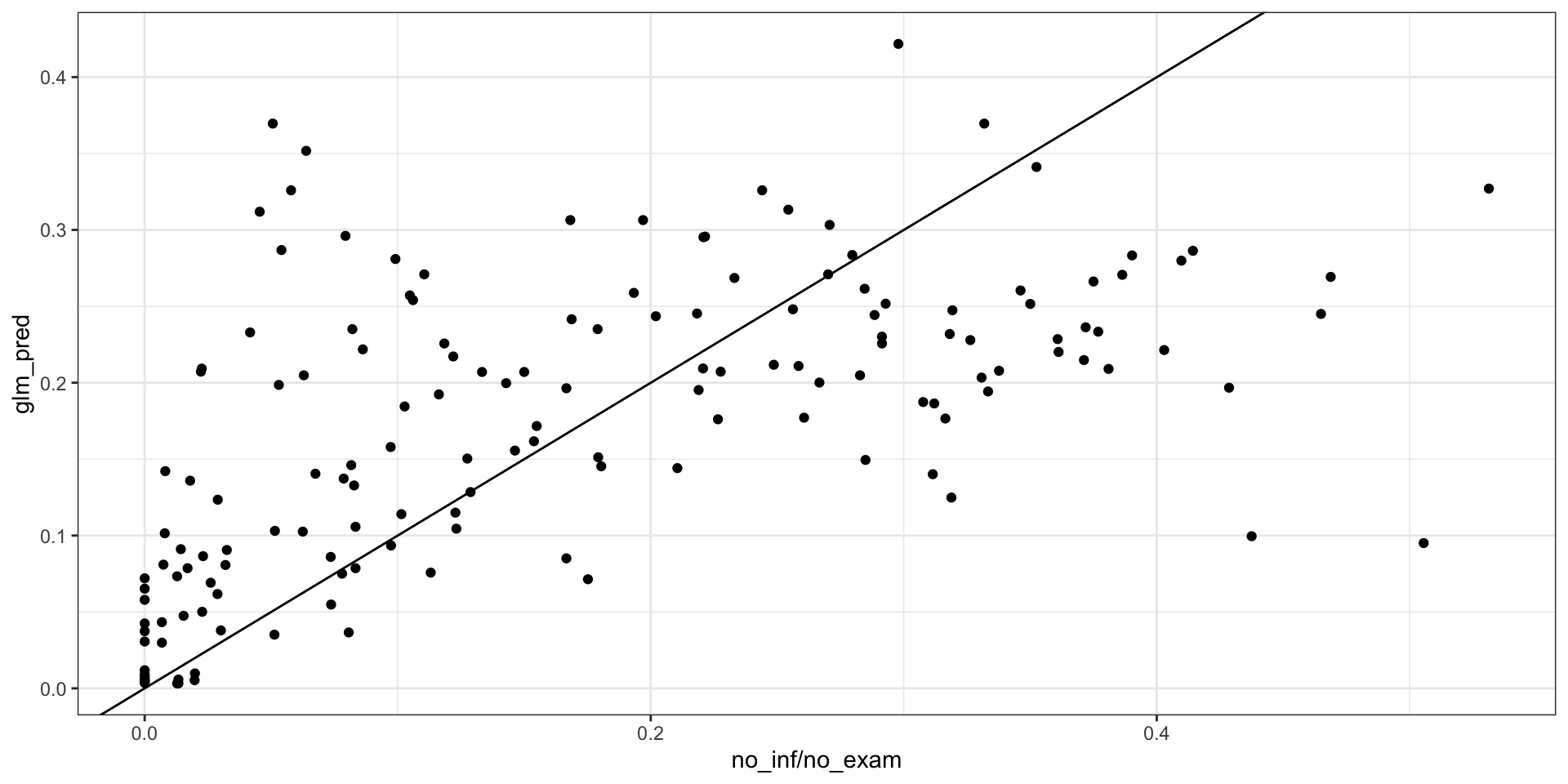
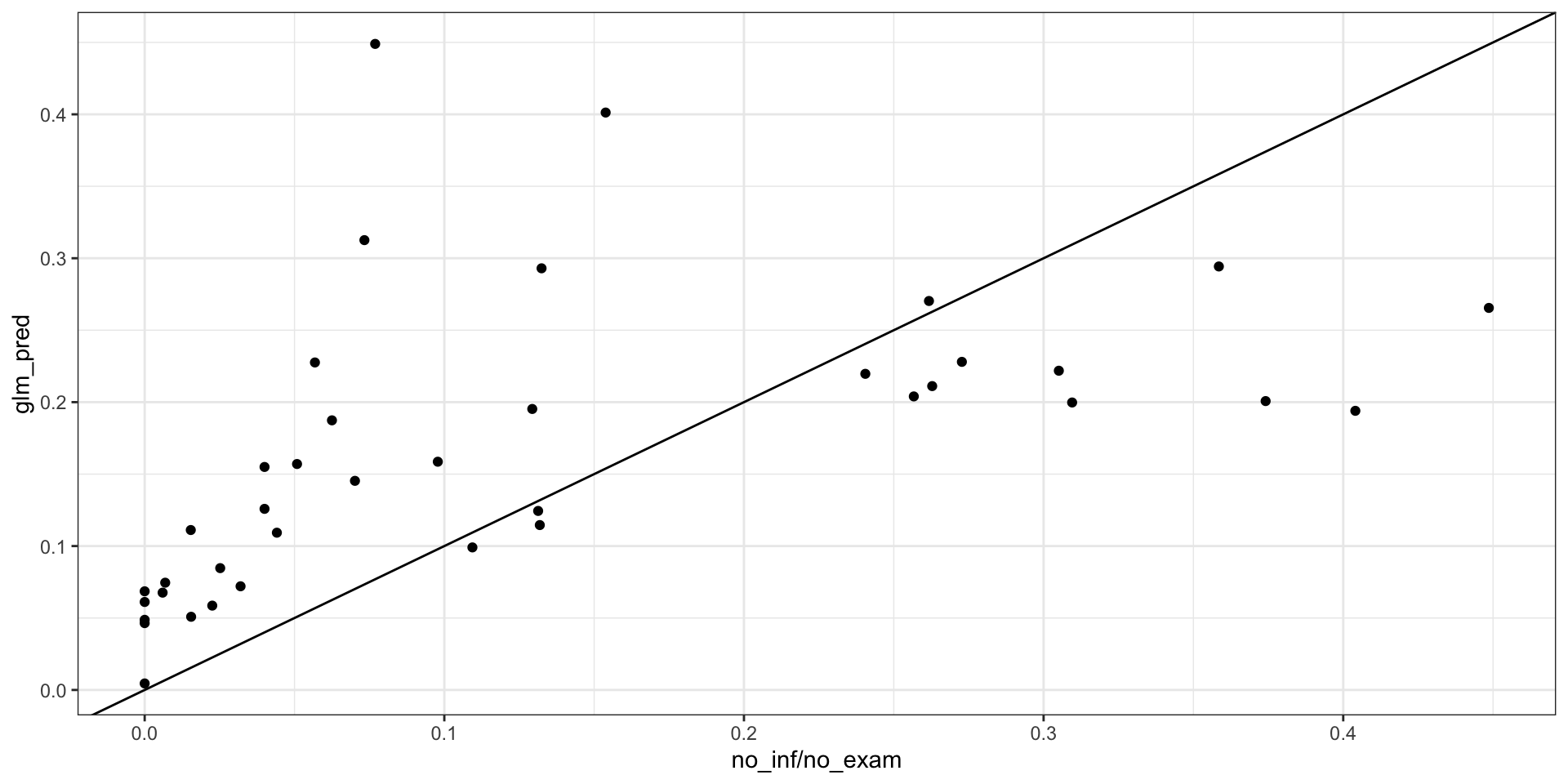
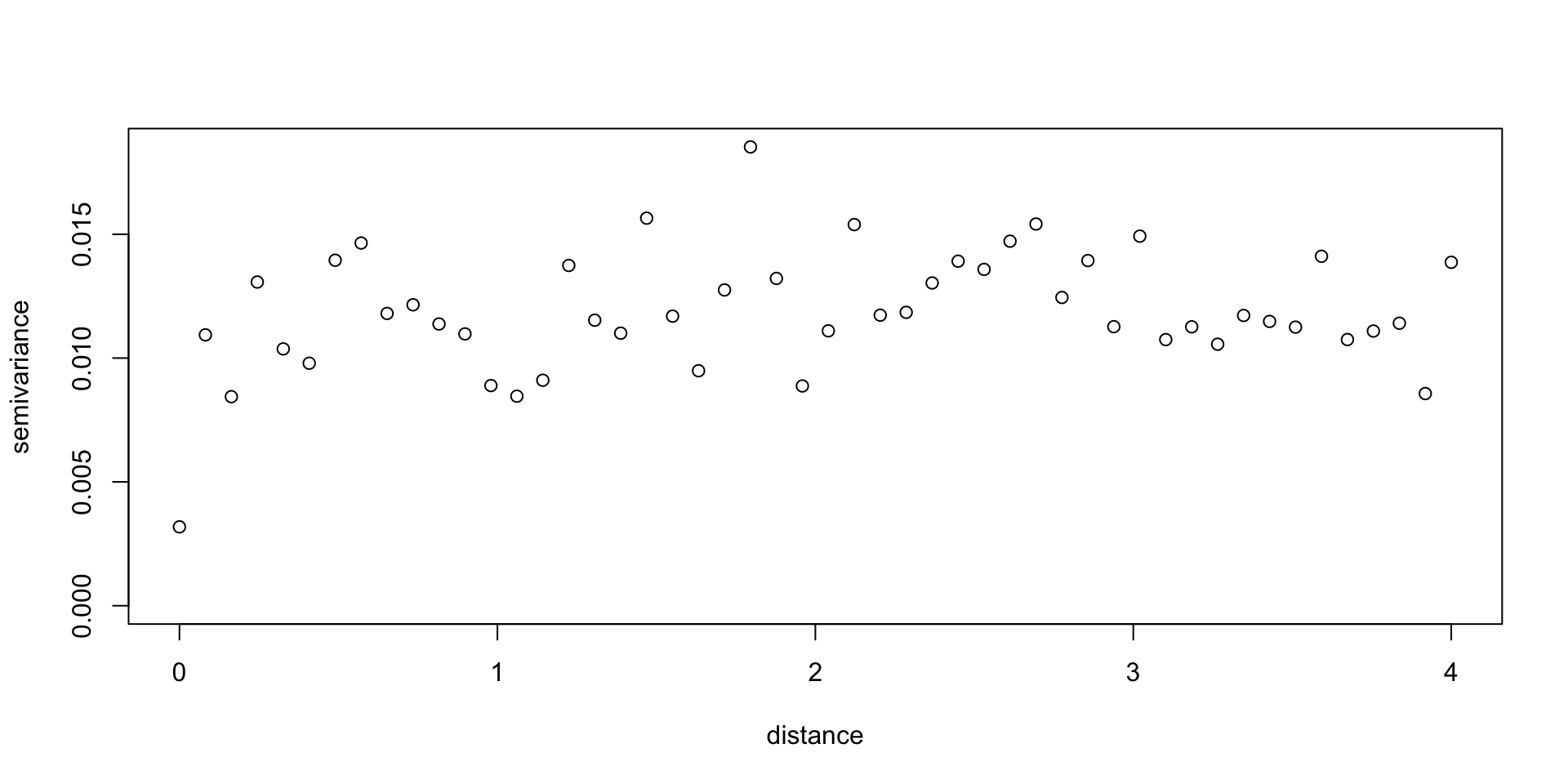
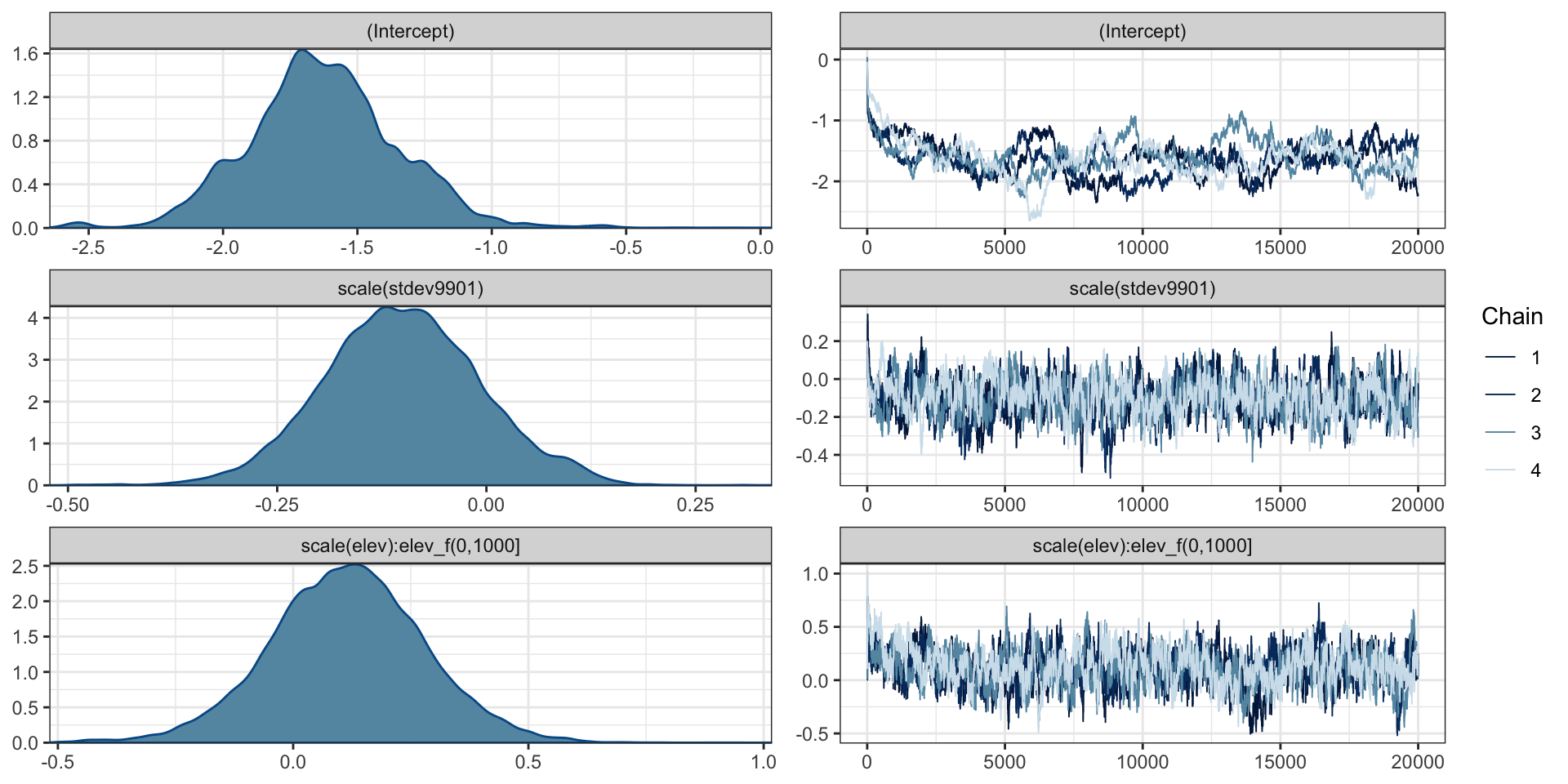
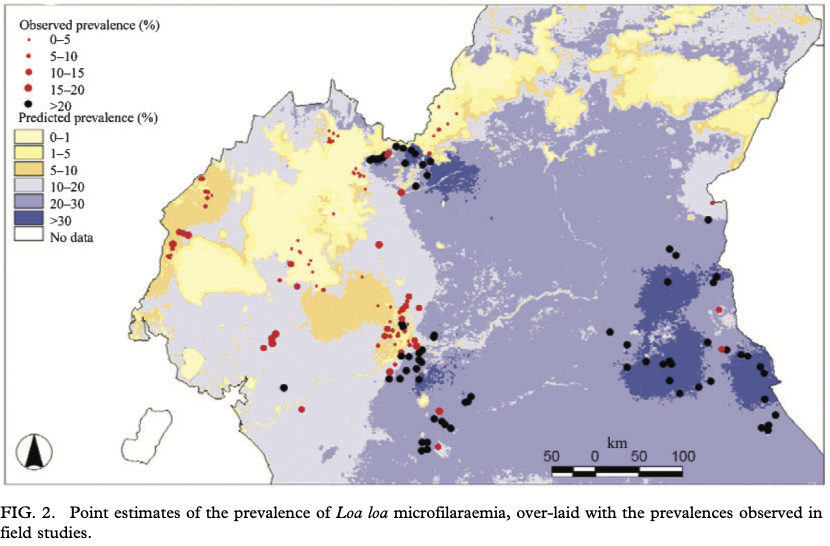
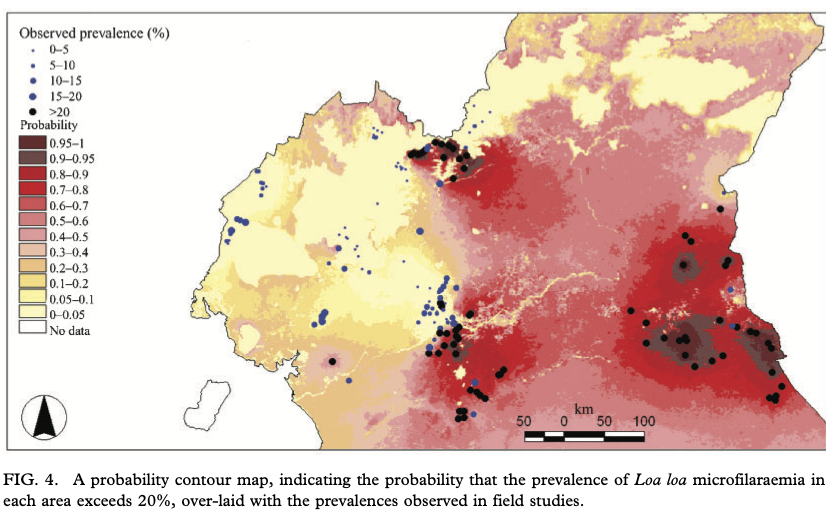
loaloa = PrevMap::loaloa %>%
as_tibble() %>%
setNames(., tolower(names(.))) %>%
rename(elev=elevation)
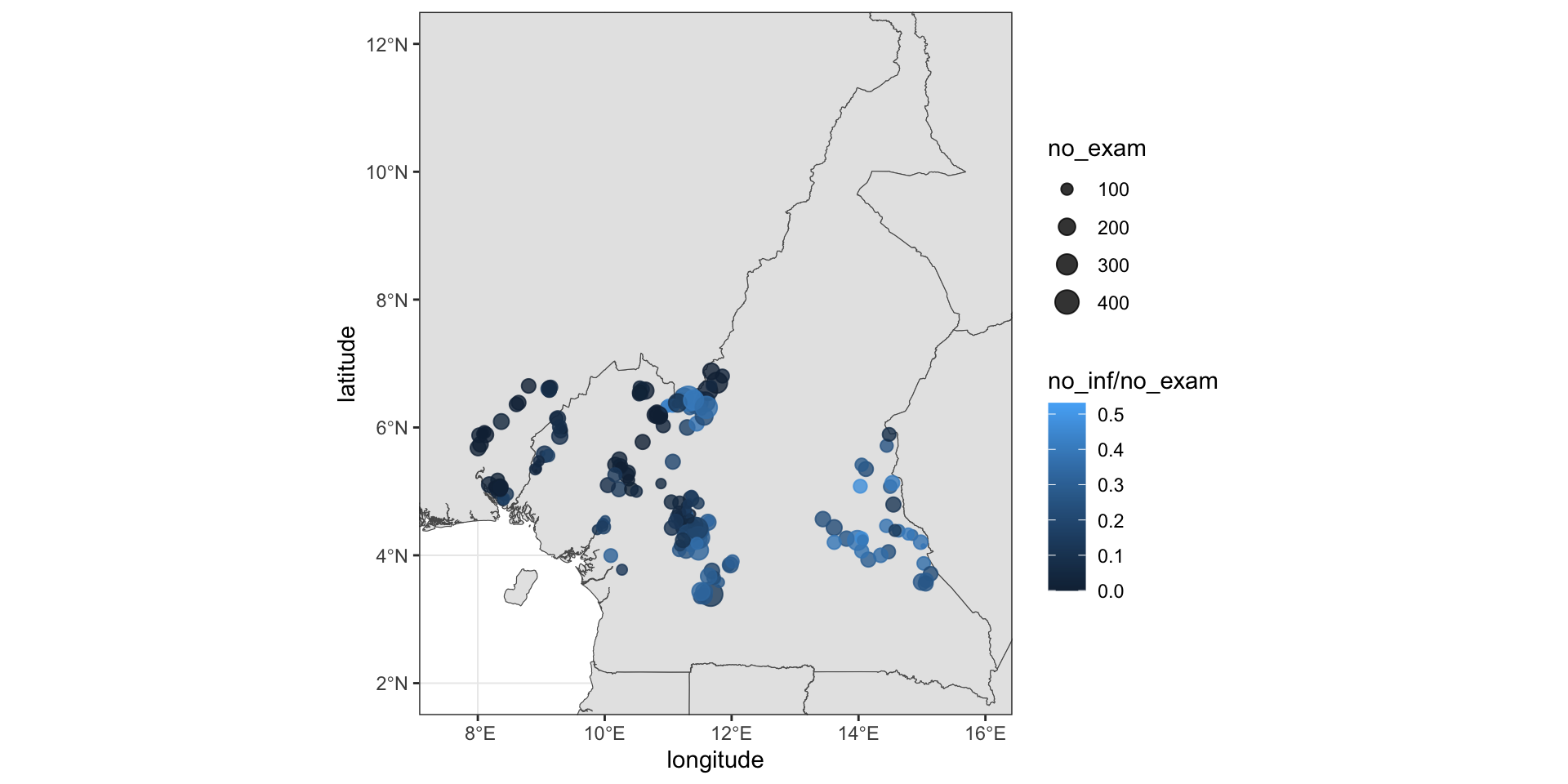
loaloa# A tibble: 197 × 11
row villcode longi…¹ latit…² no_exam no_inf elev mean9…³ max9901
<int> <int> <dbl> <dbl> <int> <int> <int> <dbl> <dbl>
1 1 214 8.04 5.74 162 0 108 0.439 0.69
2 2 215 8.00 5.68 167 1 99 0.426 0.74
3 3 118 8.91 5.35 88 5 783 0.491 0.79
4 4 219 8.10 5.92 62 5 104 0.432 0.67
5 5 212 8.18 5.10 167 3 109 0.415 0.85
6 6 116 8.93 5.36 66 3 909 0.436 0.8
7 7 16 11.4 4.88 163 11 503 0.502 0.78
8 8 217 8.07 5.90 83 0 103 0.373 0.69
9 9 112 9.02 5.59 30 4 751 0.481 0.8
10 10 104 9.31 6.00 57 4 268 0.487 0.84
# … with 187 more rows, 2 more variables: min9901 <dbl>,
# stdev9901 <dbl>, and abbreviated variable names ¹longitude,
# ²latitude, ³mean9901