[1] "avg_temp" "calc_crps" "cond_predict"
[4] "emp_semivariogram" "epred_draws_fix" "exp_cov"
[7] "exp_sv" "gpglm" "gplm"
[10] "linear_cov" "matern_cov" "normalize_weights"
[13] "nugget_cov" "periodic_cov" "pow_exp_cov"
[16] "pow_exp_sv" "predicted_draws_fix" "residual_draws_fix"
[19] "rmvnorm" "rquad_cov" "sphere_cov"
[22] "sq_exp_cov" "sq_exp_sv" "strip_attrs" Covariance Functions
Lecture 16
sta344 package
Basics
The package is based on the course organizartion and can be installed with,
collects all of the utility functions from the host scripts (e.g. util-crps.R, fix_pred_draw.R, etc.)
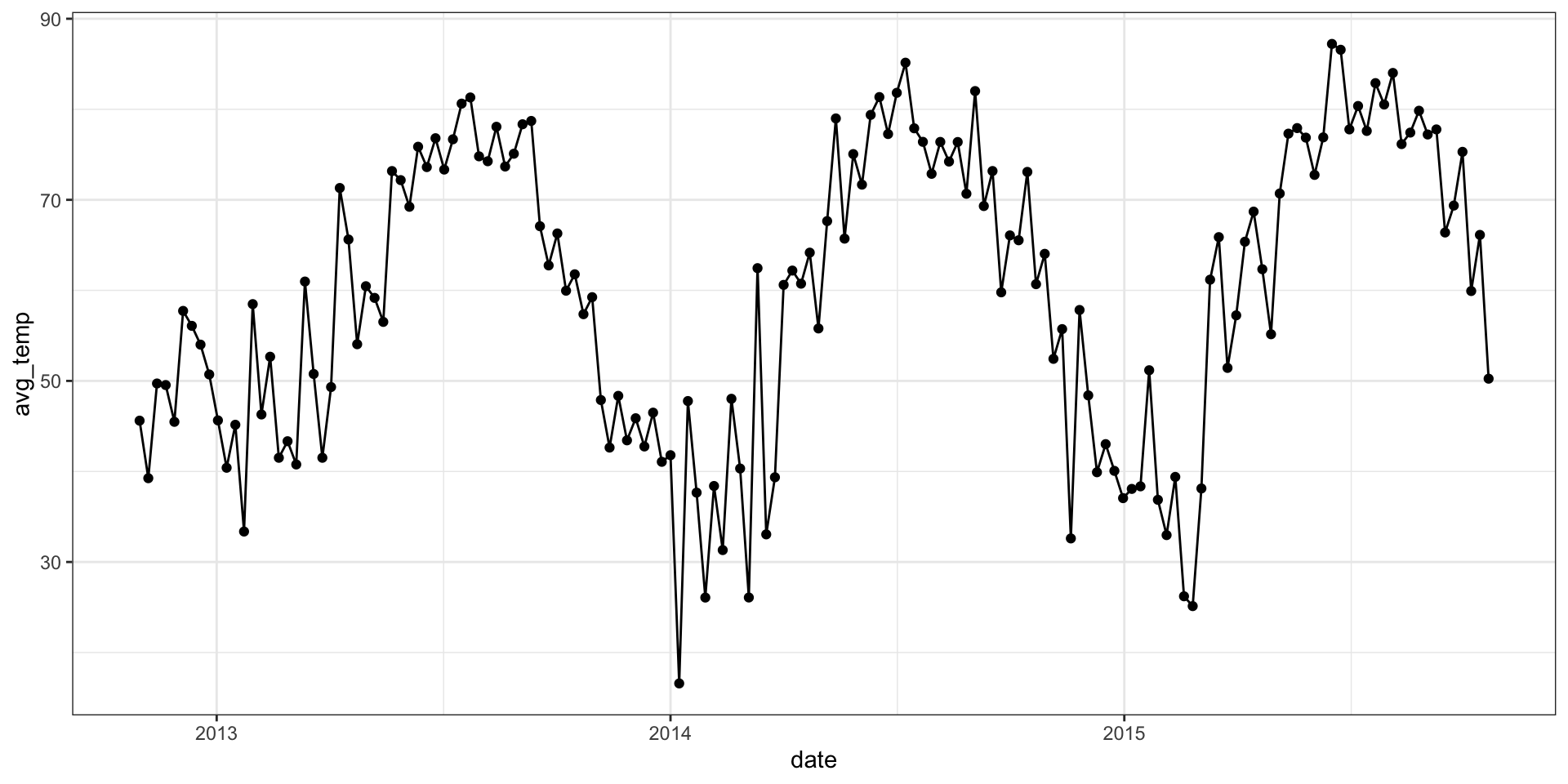
From last time
( temp = readRDS("data/avg_temp_df.rds") %>%
slice(1:(52*3)) %>%
mutate(week = as.numeric(date - date[1])/7) )# A tibble: 156 × 3
date avg_temp week
<date> <dbl> <dbl>
1 2012-10-31 45.6 0
2 2012-11-07 39.3 1
3 2012-11-14 49.7 2
4 2012-11-21 49.5 3
5 2012-11-28 45.5 4
6 2012-12-05 57.7 5
7 2012-12-12 56.1 6
8 2012-12-19 54.0 7
9 2012-12-26 50.7 8
10 2013-01-02 45.6 9
# … with 146 more rowsEmpirical semi-variogram
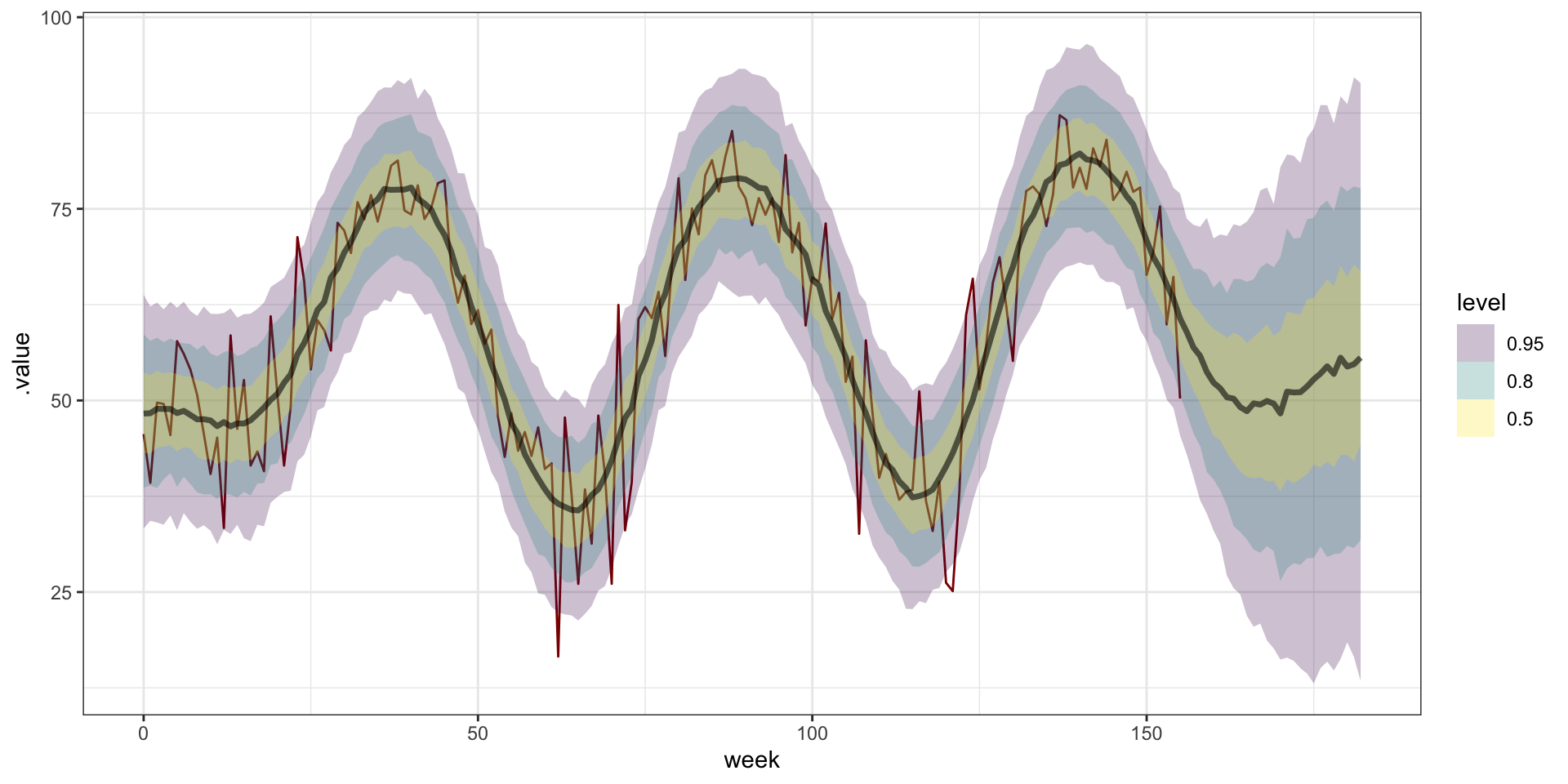
Model fitting
m = gplm(
avg_temp~1, data = temp, coords = "week",
cov_model = "gaussian",
starting=list(
"phi"=sqrt(3)/4, "sigma.sq"=1, "tau.sq"=1
),
tuning=list(
"phi"=1, "sigma.sq"=1, "tau.sq"=1
),
priors=list(
"phi.unif"=c(sqrt(3)/52, sqrt(3)/1),
"sigma.sq.ig"=c(2, 1),
"tau.sq.ig"=c(2, 1)
),
thin = 5,
n_batch = 100,
batch_len = 100
)Model plots
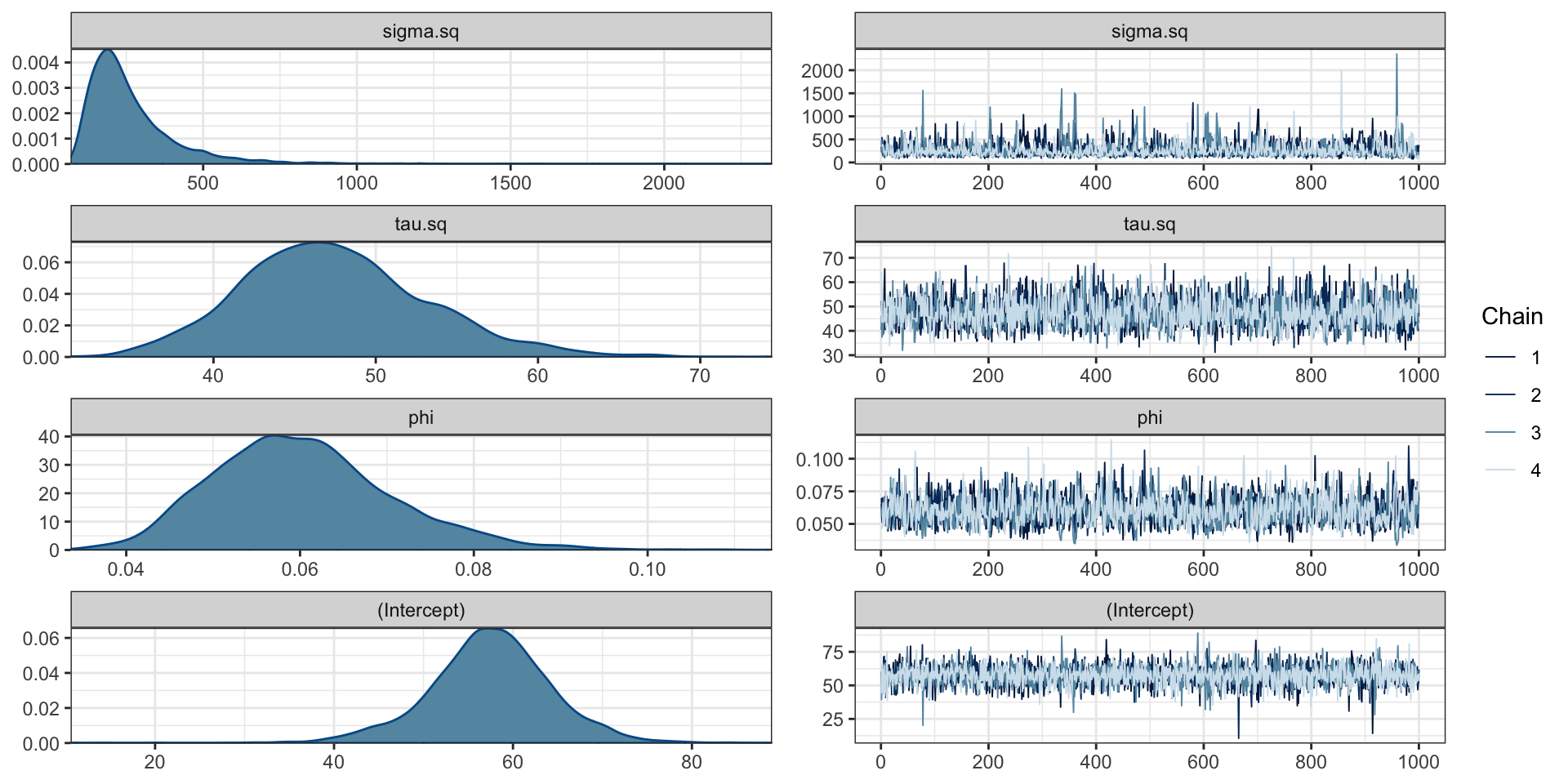
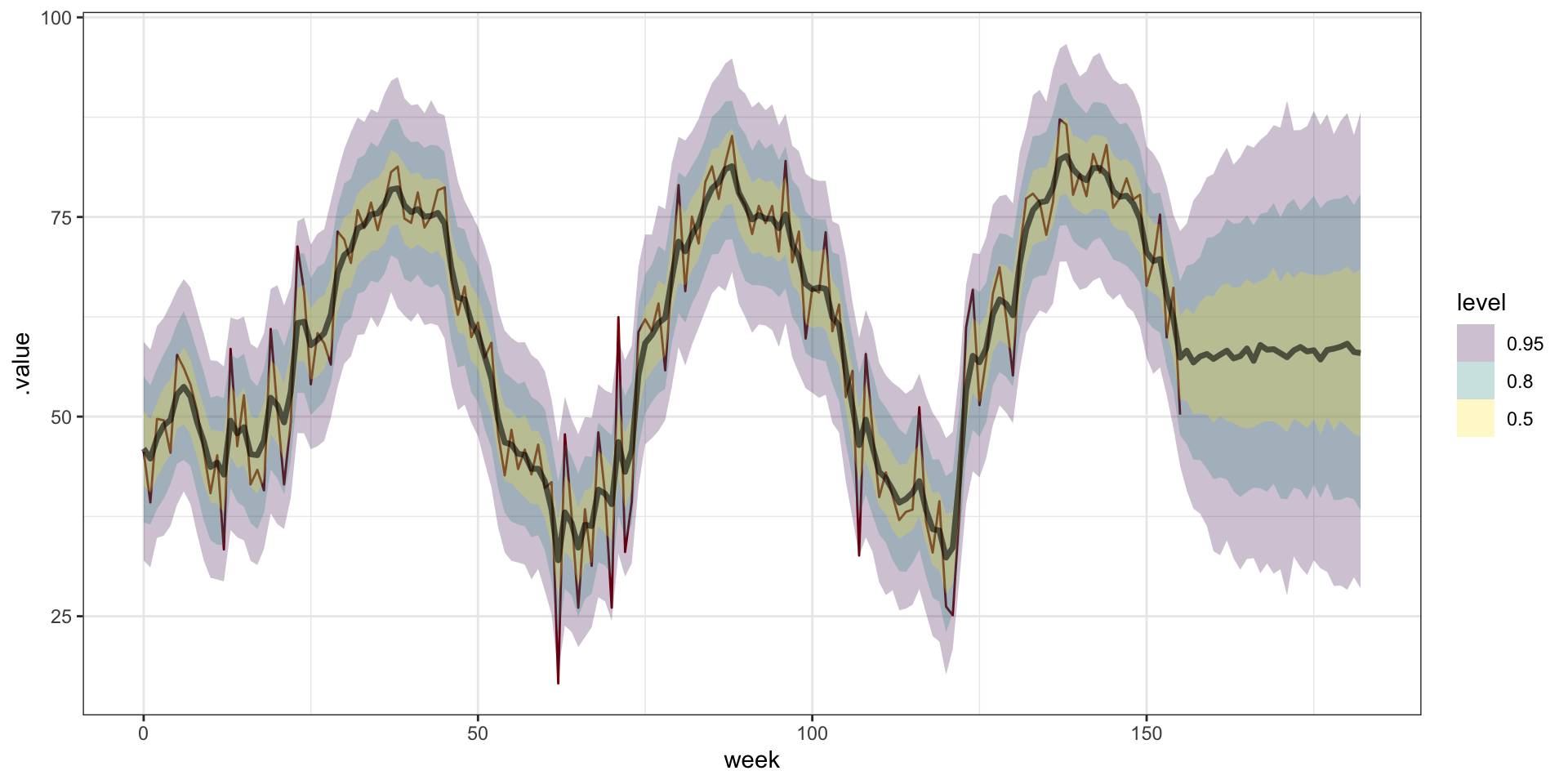
Prediction
newdata = data.frame(
week = seq(0,3.5*52) |> jitter()
)
(p = predict(m, newdata=newdata, coords = "week"))# A draws_matrix: 1000 iterations, 4 chains, and 183 variables
variable
draw y[1] y[2] y[3] y[4] y[5] y[6] y[7] y[8]
1 38 45 57 42 43 47 43 55
2 51 54 47 47 46 36 50 46
3 42 56 45 57 36 54 48 60
4 48 43 52 39 53 48 54 59
5 39 38 39 46 50 50 49 48
6 65 43 50 45 48 43 55 57
7 43 52 56 60 54 71 40 58
8 51 35 55 48 54 45 45 49
9 50 48 42 44 38 52 58 40
10 38 44 33 49 41 49 48 54
# ... with 3990 more draws, and 175 more variablesResults
Exponential covariance
m_exp = gplm(
avg_temp~1, data = temp, coords = "week",
cov_model = "exponential",
starting=list(
"phi"=3/4, "sigma.sq"=1, "tau.sq"=1
),
tuning=list(
"phi"=1, "sigma.sq"=1, "tau.sq"=1
),
priors=list(
"phi.unif"=c(3/52, 3/1),
"sigma.sq.ig"=c(2, 1),
"tau.sq.ig"=c(2, 1)
),
thin = 5,
n_batch = 100,
batch_len = 100
)
p_exp = predict(m_exp, newdata=newdata, coords = "week")Results
More Covariance Functions
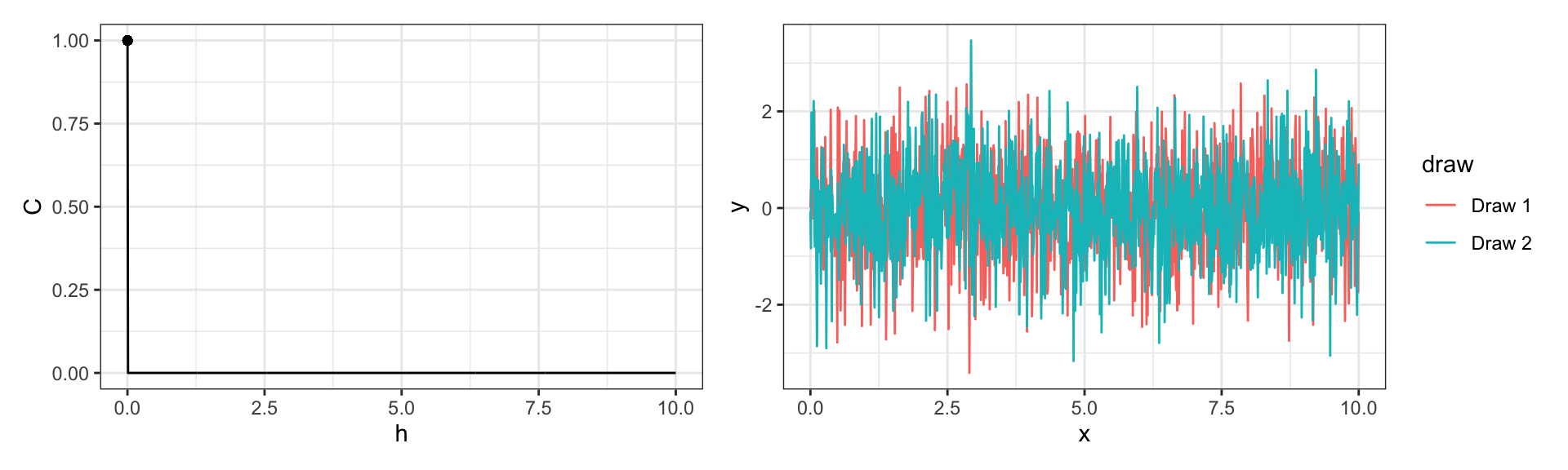
Nugget Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \sigma^2 {1}_{\{h=0\}} \text{ where } h = |t_i - t_j|\]
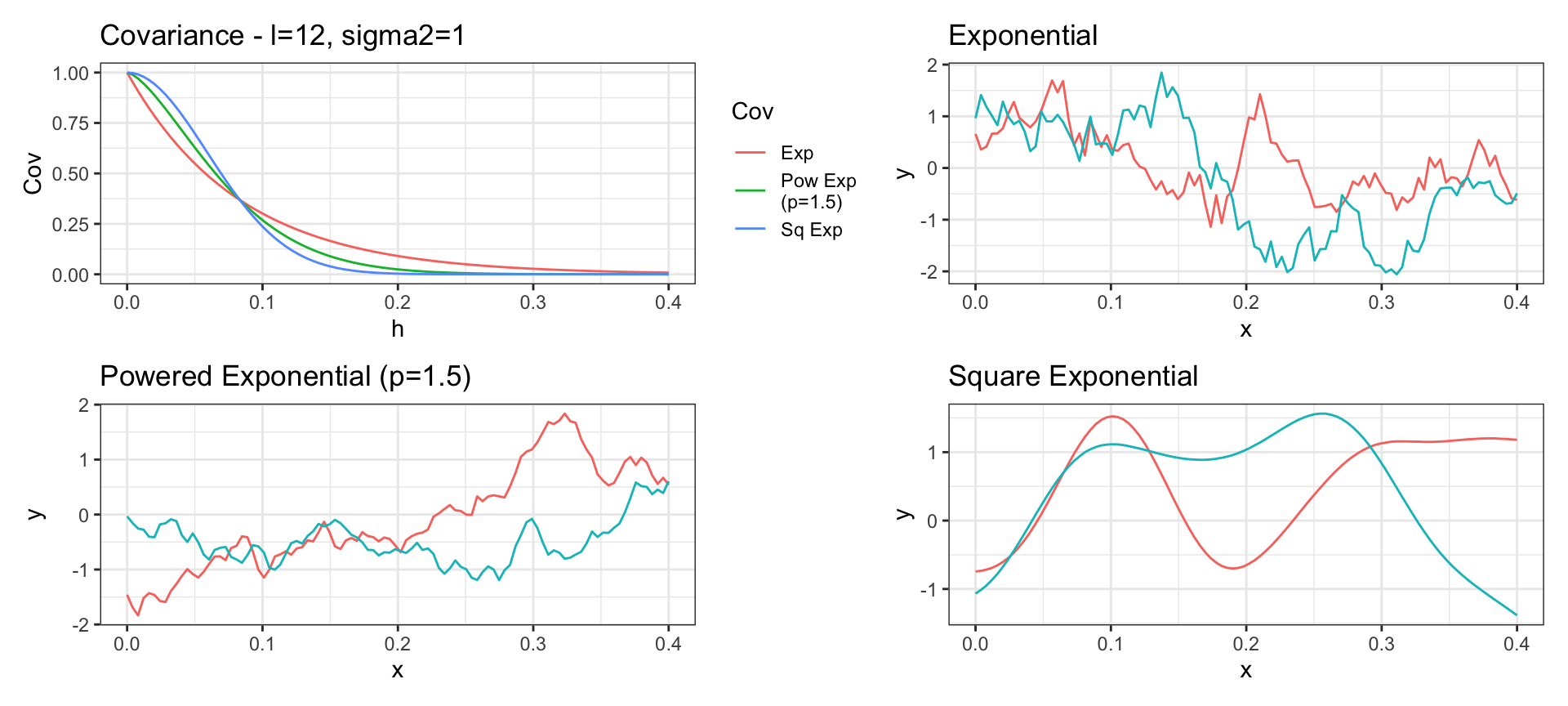
(- / Power / Square) Exponential Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \sigma^2\exp\left(-(h\,l)^p\right) \text{ where } h = |t_i - t_j|\]
Matern Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \sigma^2 ~ \frac{2^{1-\nu}}{\Gamma(\nu)} ~ \left(\sqrt{2\nu}\, h \cdot l\right)^\nu ~ K_\nu\left(\sqrt{2\nu} \, h \cdot l\right) \text{ where } h = |t_i - t_j|\]
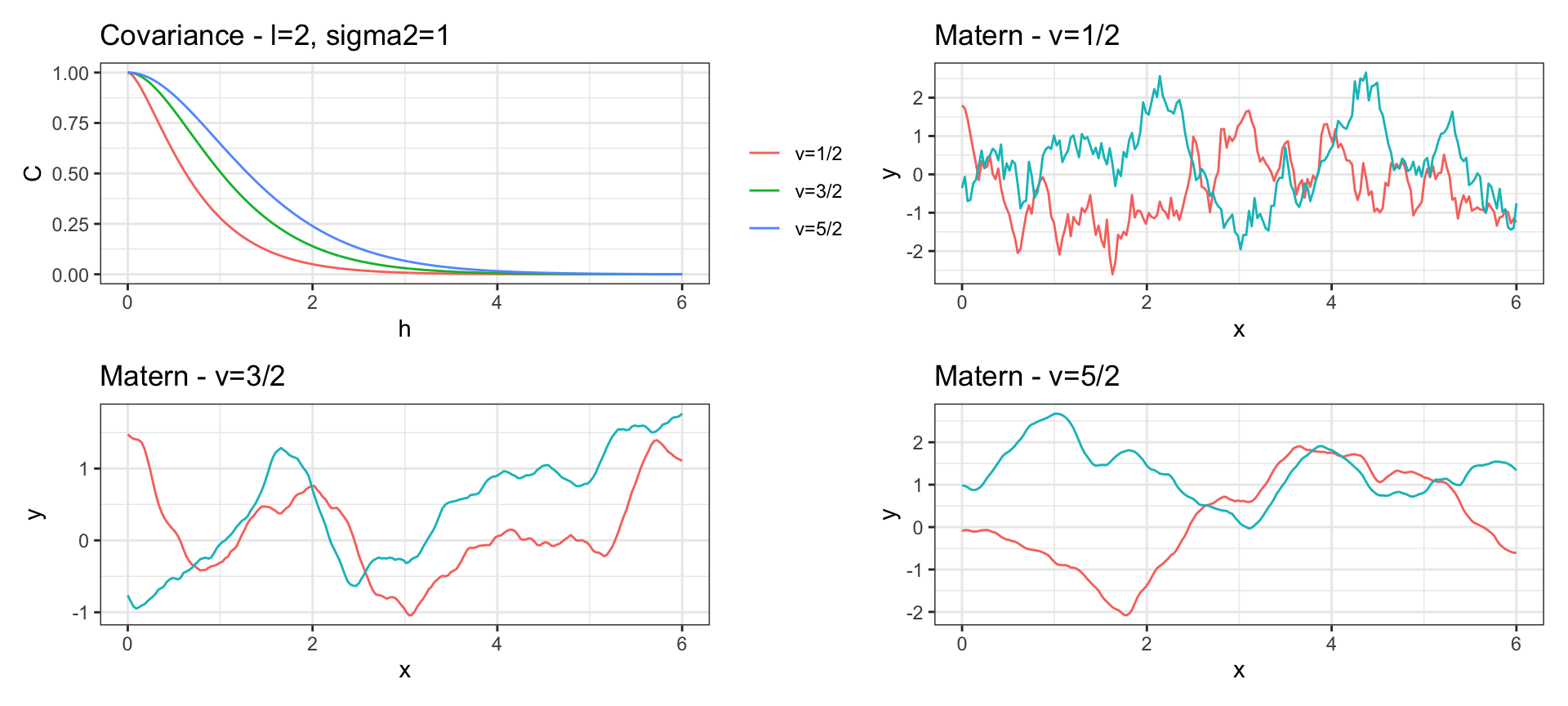
Matern Covariance
\(K_\nu()\) is the modified Bessel function of the second kind.
A Gaussian process with Matérn covariance has sample functions that are \(\lceil \nu -1\rceil\) times differentiable.
When \(\nu = 1/2 + p\) for \(p \in \mathbb{N}^+\) then the Matern has a simplified form
When \(\nu = 1/2\) the Matern is equivalent to the exponential covariance.
As \(\nu \to \infty\) the Matern converges to the squared exponential covariance.
Rational Quadratic Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \sigma^2 \left(1 + \frac{h^2 \, l^2}{\alpha}\right)^{-\alpha} \text{ where } h = |t_i - t_j|\]
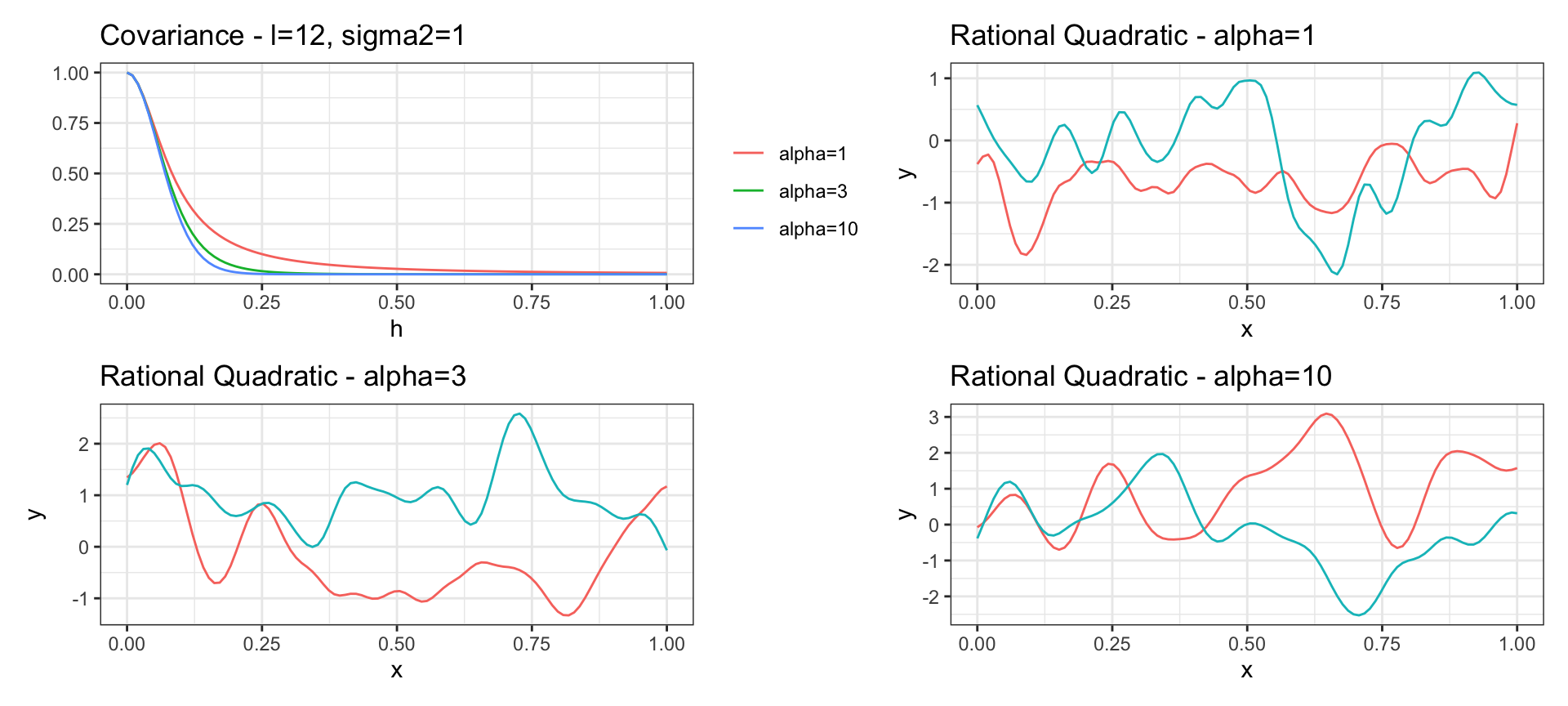
Rational Quadratic Covariance
is a scaled mixture of squared exponential covariance functions with different characteristic length-scales (\(l\)).
As \(\alpha \to \infty\) the rational quadratic converges to the square exponential covariance.
Has sample functions that are infinitely differentiable for any value of \(\alpha\)
Spherical Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \begin{cases} \sigma^2\left(1 - \frac{3}{2} h \cdot l + \frac{1}{2} (h \cdot l)^3)\right) & \text{if } 0 < h < 1/l \\ 0 & \text{otherwise} \end{cases} \text{ where } h = |t_i - t_j|\]
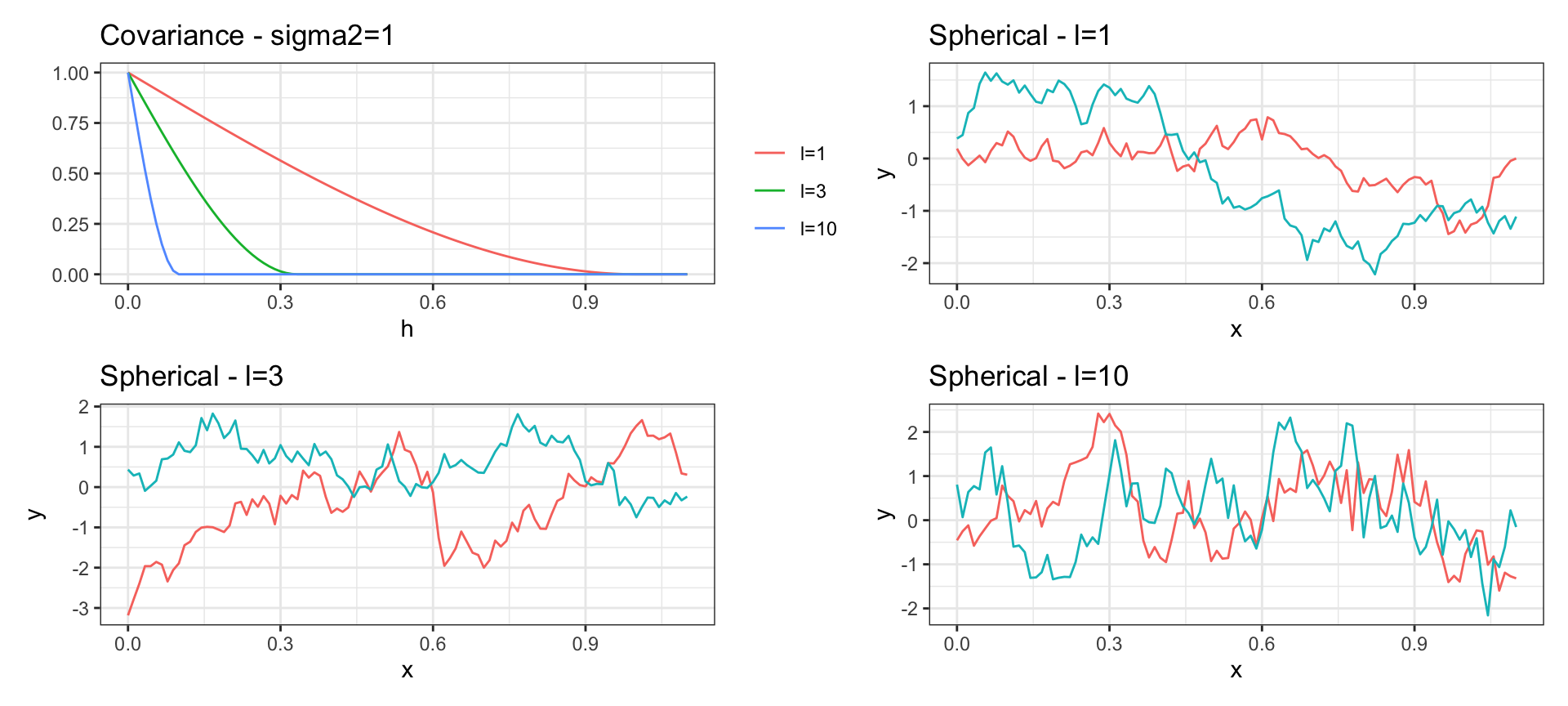
Periodic Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \sigma^2 \exp\left(-2\, l^2 \sin^2\left(\pi\frac{h}{p}\right)\right) \text{ where } h = |t_i - t_j| \]
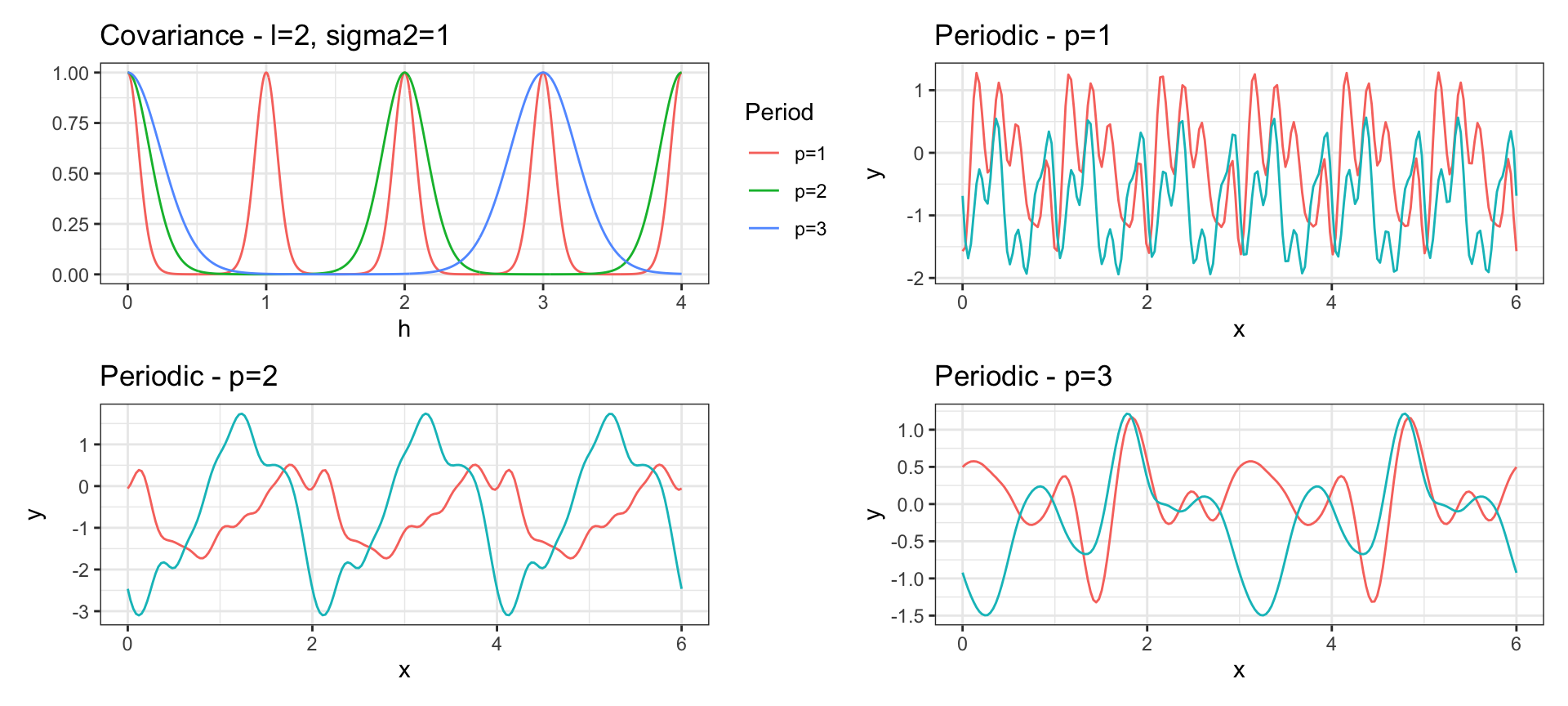
Linear Covariance
\[ Cov(y_{t_i}, y_{t_j}) = \sigma^2_b + \sigma^2_v~(t_i-c)(t_j-c)\]
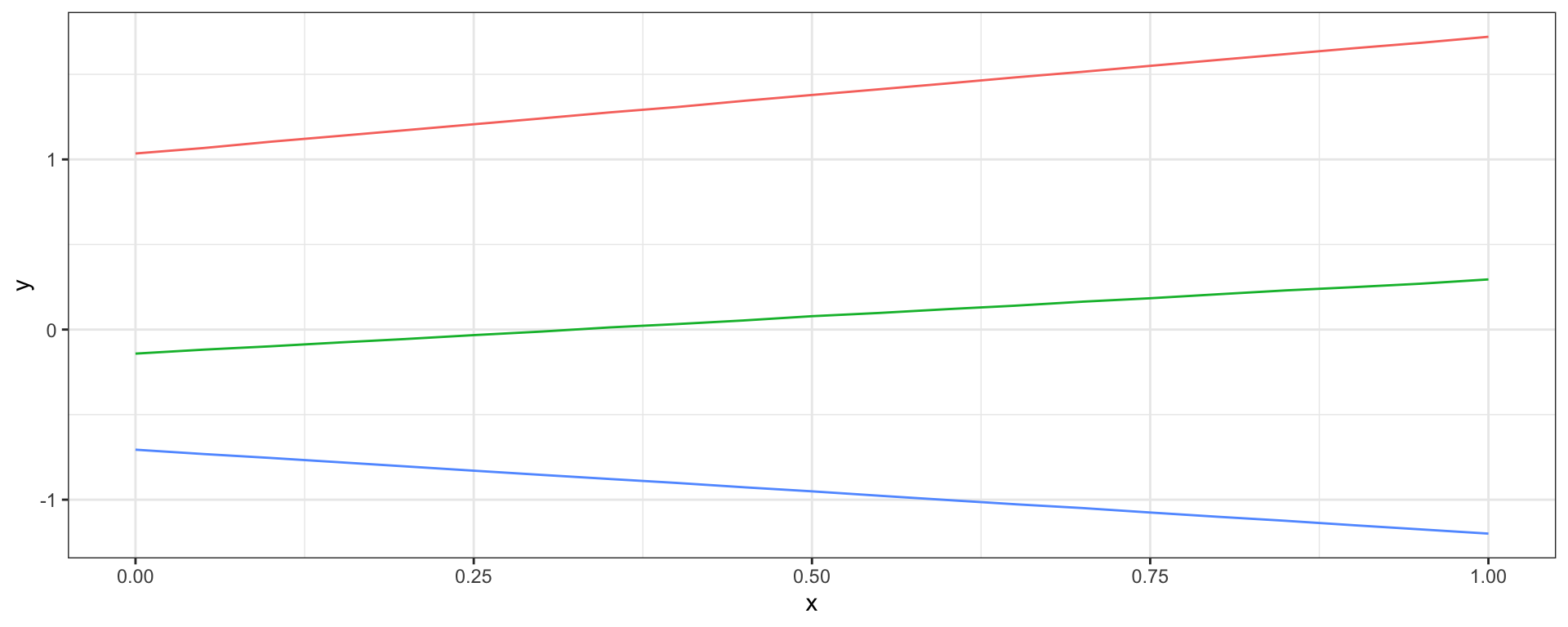
Combining Covariances
If we definite two valid covariance functions, \(Cov_a(y_{t_i}, y_{t_j})\) and \(Cov_b(y_{t_i}, y_{t_j})\) then the following are also valid covariance functions,
\[ \begin{aligned} Cov_a(y_{t_i}, y_{t_j}) + Cov_b(y_{t_i}, y_{t_j}) \\ \\ Cov_a(y_{t_i}, y_{t_j}) \times Cov_b(y_{t_i}, y_{t_j}) \end{aligned} \]
Linear \(\times\) Linear \(\to\) Quadratic
\[ Cov_a(y_{t_i}, y_{t_j}) = 1 + 2~(t_i \times t_j) \] \[ Cov_b(y_{t_i}, y_{t_j}) = 2 + 1~(t_i \times t_j) \]
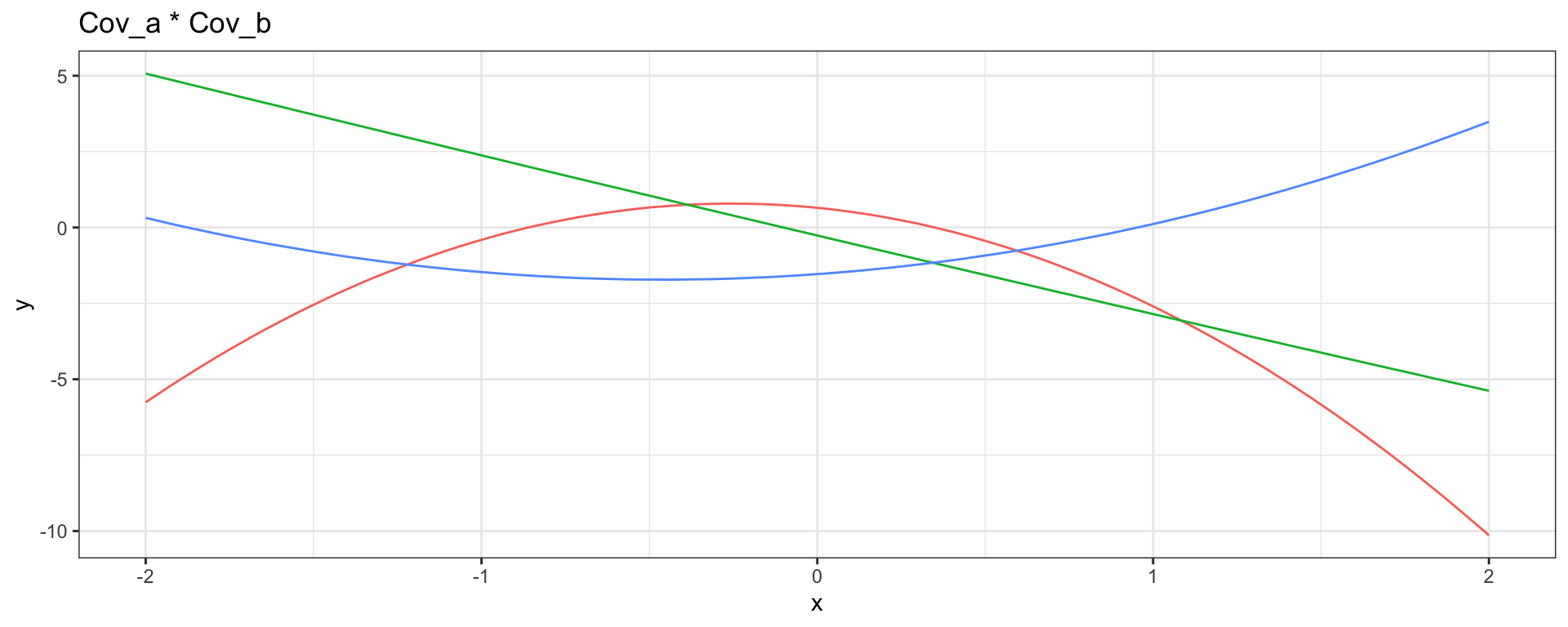
Linear \(\times\) Periodic
\[ Cov_a(y_{t_i}, y_{t_j}) = 1 + 1~(t_i \times t_j) \] \[ Cov_b(y_{t_i}, y_{t_j}) = \exp\left(-2\, \sin^2\left(2\pi\,h\right)\right) \]
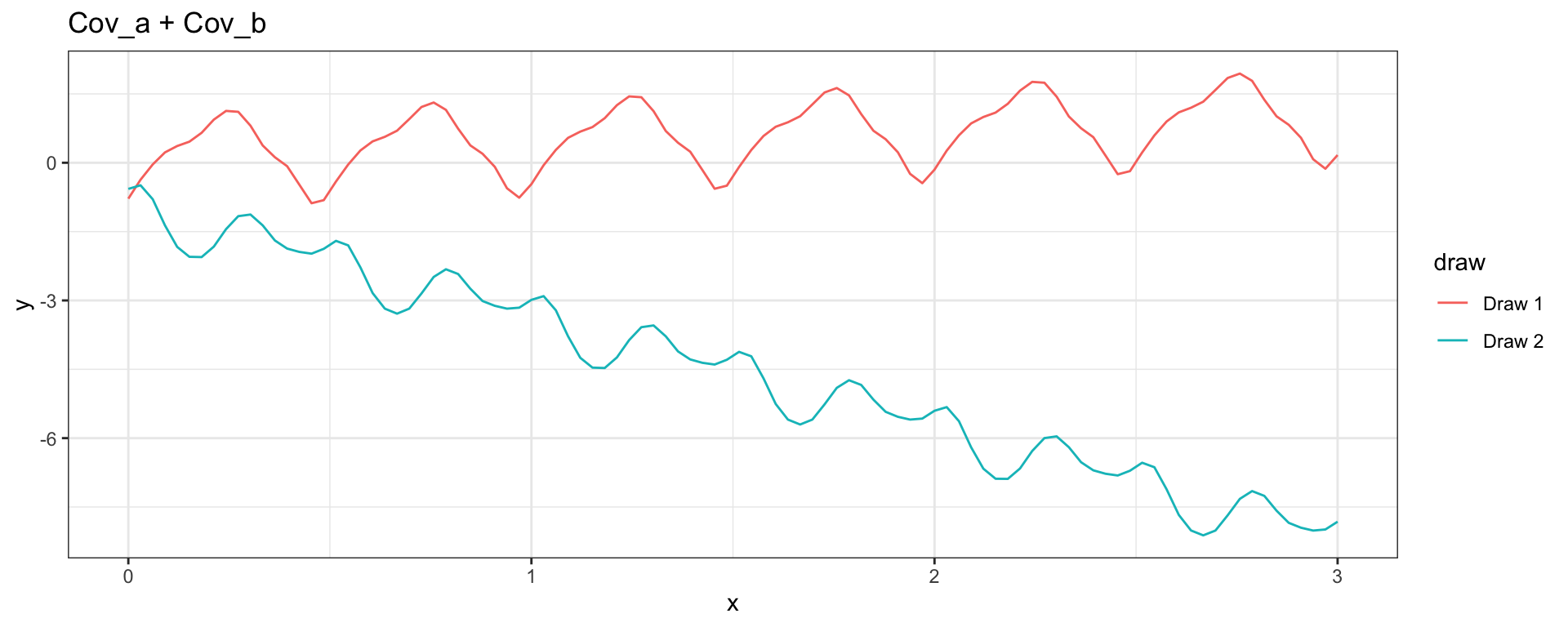
Linear + Periodic
\[ Cov_a(y_{t_i}, y_{t_j}) = 1 + 1~(t_i \times t_j) \] \[ Cov_b(h = |t_i - t_j|) = \exp\left(-2\, \sin^2\left(2\pi\,h\right)\right) \]
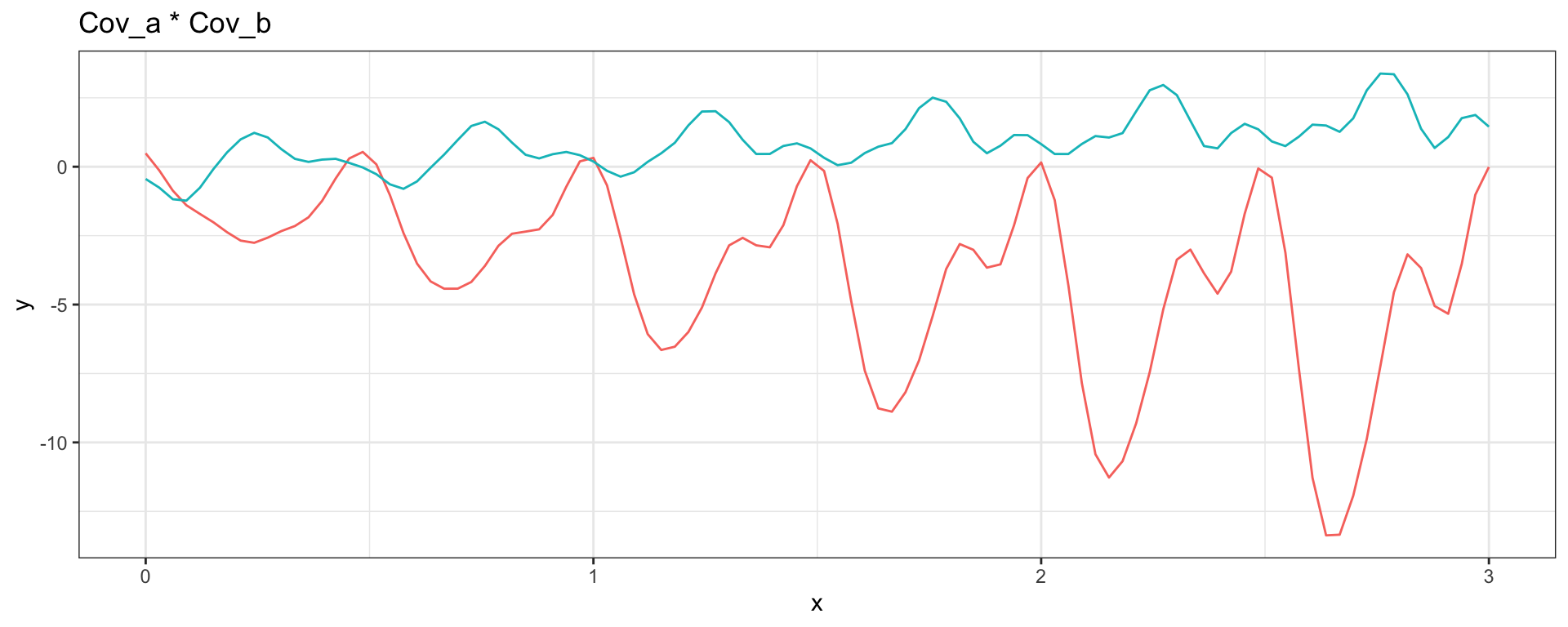
Sq Exp \(\times\) Periodic \(\to\) Locally Periodic
\[ Cov_a(h = |t_i - t_j|) =\exp(-(1/3)h^2) \] \[ Cov_b(h = |t_i - t_j|) = \exp\left(-2\, \sin^2\left(\pi\,h\right)\right) \]
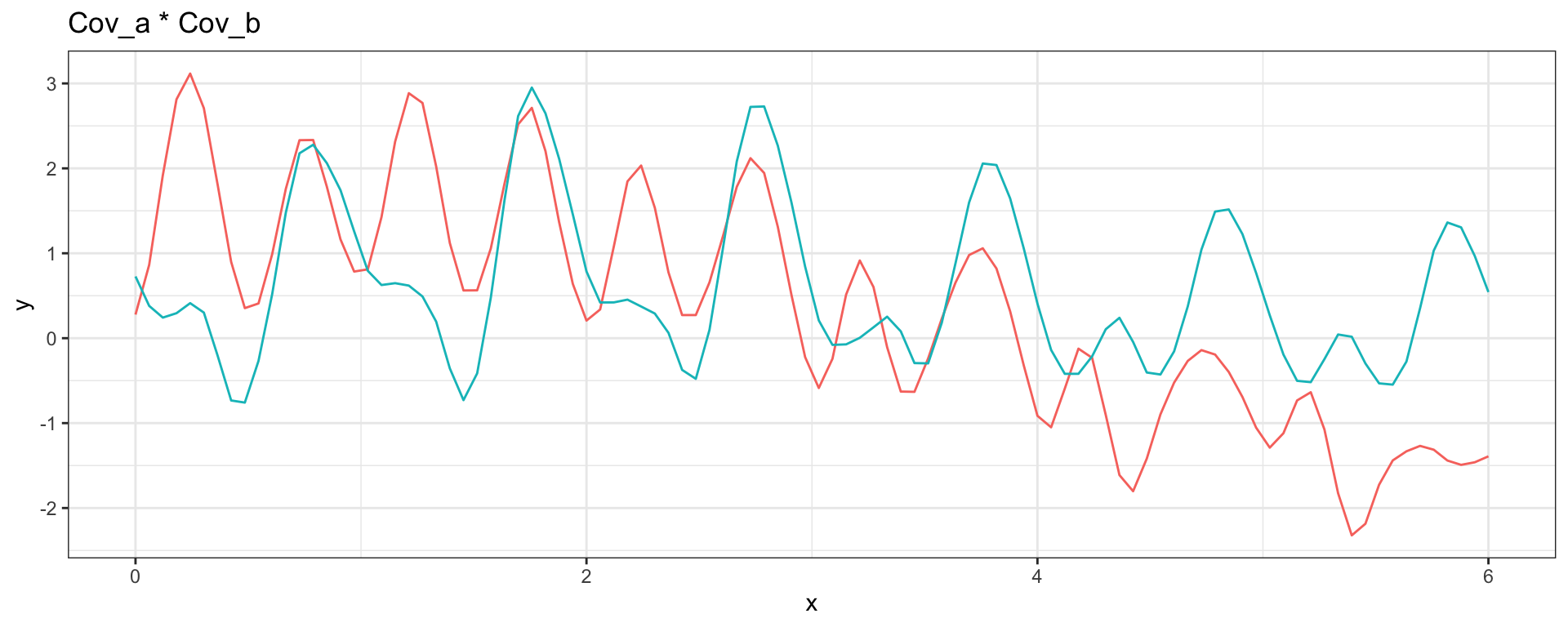
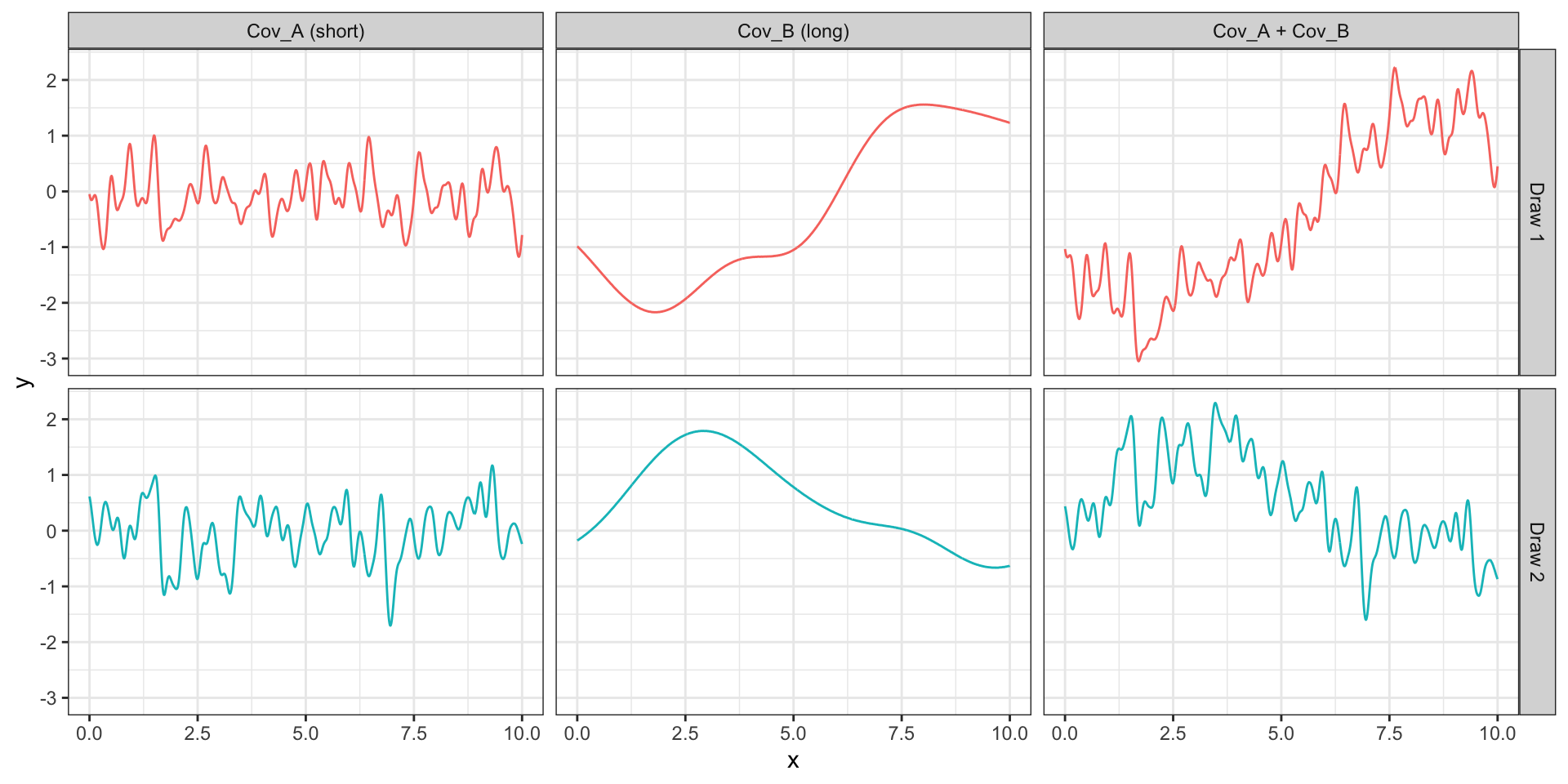
Sq Exp (short) + Sq Exp (long)
\[ Cov_a(h = |t_i - t_j|) = (1/4) \exp(-4\sqrt{3}h^2) \] \[ Cov_b(h = |t_i - t_j|) = \exp(-(\sqrt{3}/2)h^2) \]
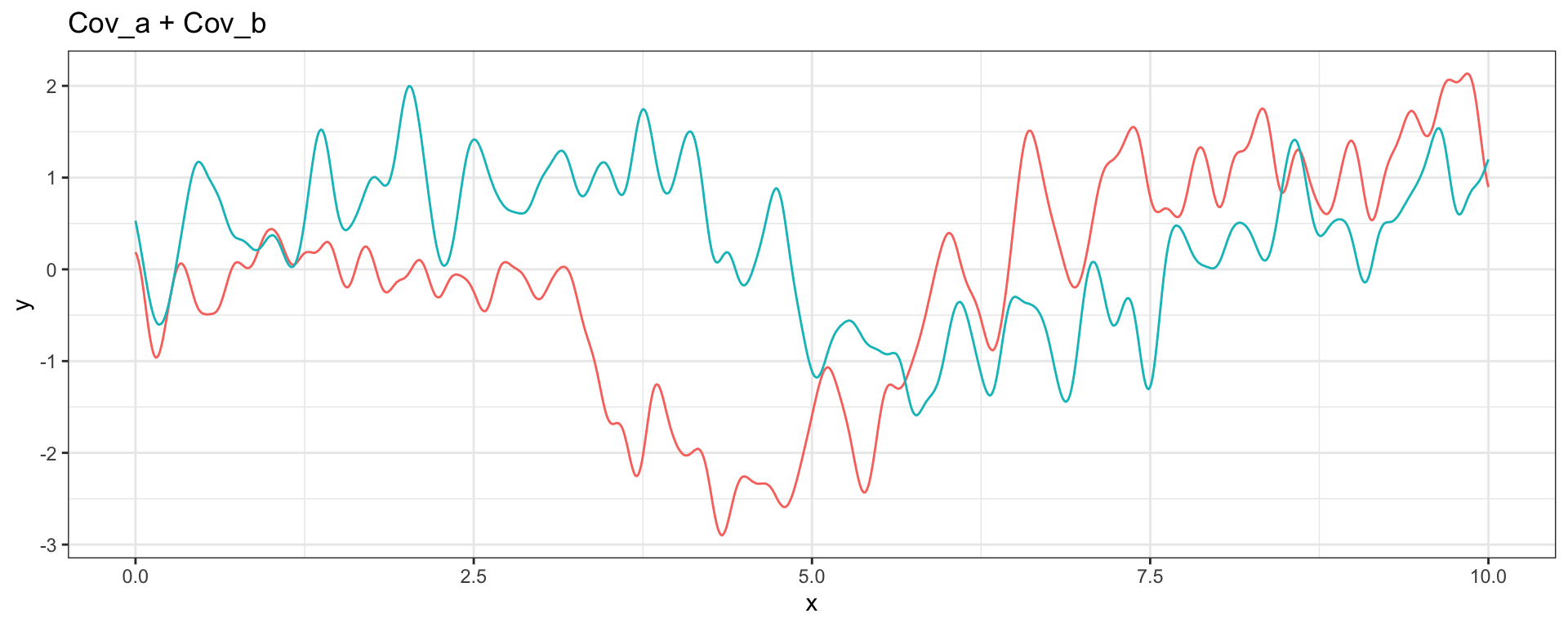
Seen another way
BDA3 example
BDA3
Births (one year)
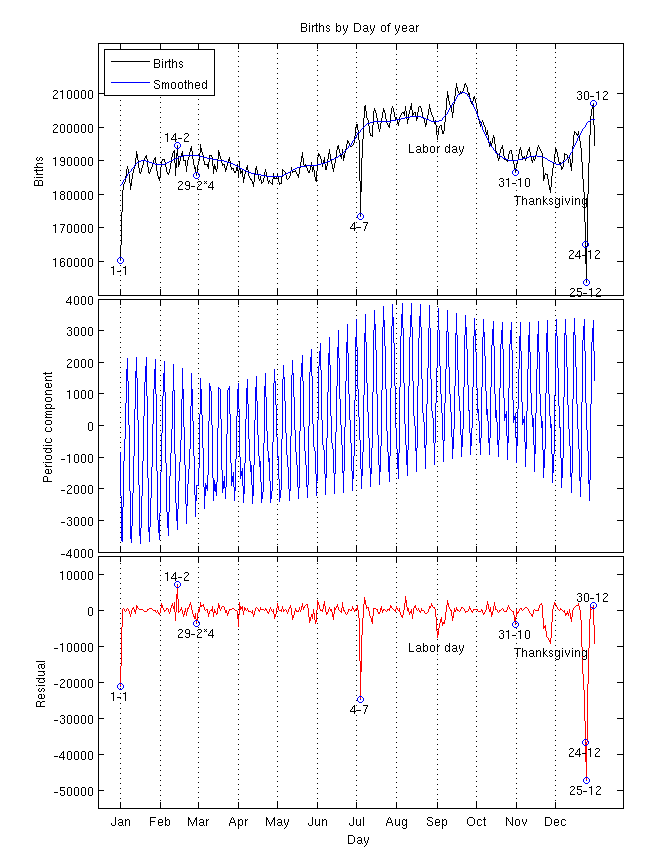
Smooth long term trend
(sq exp cov)Seven day periodic trend with decay (periodic x sq exp cov)
Constant mean
Component Contributions
We can view our GP in the following ways (marginal form),
\[ \boldsymbol{y} \sim N(\boldsymbol{\mu},~ \boldsymbol{\Sigma}_1 + \boldsymbol{\Sigma}_2 + \sigma^2 \boldsymbol{I}\,) \]
but with appropriate conditioning we can also think of \(\boldsymbol{y}\) as being the sum of multiple independent GPs (latent form)
\[ \boldsymbol{y} = \boldsymbol{\mu} + w_1(\boldsymbol{t}) + w_2(\boldsymbol{t}) + w_3(\boldsymbol{t})\] where \[ \begin{aligned} w_1(\boldsymbol{t}) &\sim N(0, \boldsymbol{\Sigma}_1) \\ w_2(\boldsymbol{t}) &\sim N(0, \boldsymbol{\Sigma}_2) \\ w_3(\boldsymbol{t}) &\sim N(0, \sigma^2 \boldsymbol{I}\,) \end{aligned} \]
Decomposition of Covariance Components
\[ \begin{bmatrix} y \\ w_1 \\ w_2 \end{bmatrix} \sim N \left( \begin{bmatrix} \boldsymbol{\mu} \\ 0 \\ 0 \end{bmatrix},~ \begin{bmatrix} \Sigma_1 + \Sigma_2 + \sigma^2 \boldsymbol{I} & \Sigma_1 & \Sigma_2 \\ \Sigma_1 & \Sigma_1 & 0\\ \Sigma_2 & 0 & \Sigma_2 \\ \end{bmatrix} \right) \]
therefore
\[ w_1 ~|~ \boldsymbol{y},\boldsymbol{\mu},\boldsymbol{\theta} \sim N(\boldsymbol{\mu}_{cond},~ \boldsymbol{\Sigma}_{cond}) \]
\[ \boldsymbol{\mu}_{cond} = 0 + \Sigma_1 ~ (\Sigma_1 + \Sigma_2 + \sigma^2 I)^{-1}(\boldsymbol{y}-\boldsymbol{\mu}) \] \[ \boldsymbol{\Sigma}_{cond} = \Sigma_1 - \Sigma_1 (\Sigma_1 + \Sigma_2 + \sigma^2 \boldsymbol{I})^{-1} {\Sigma_1}^t \]
Births (multiple years)
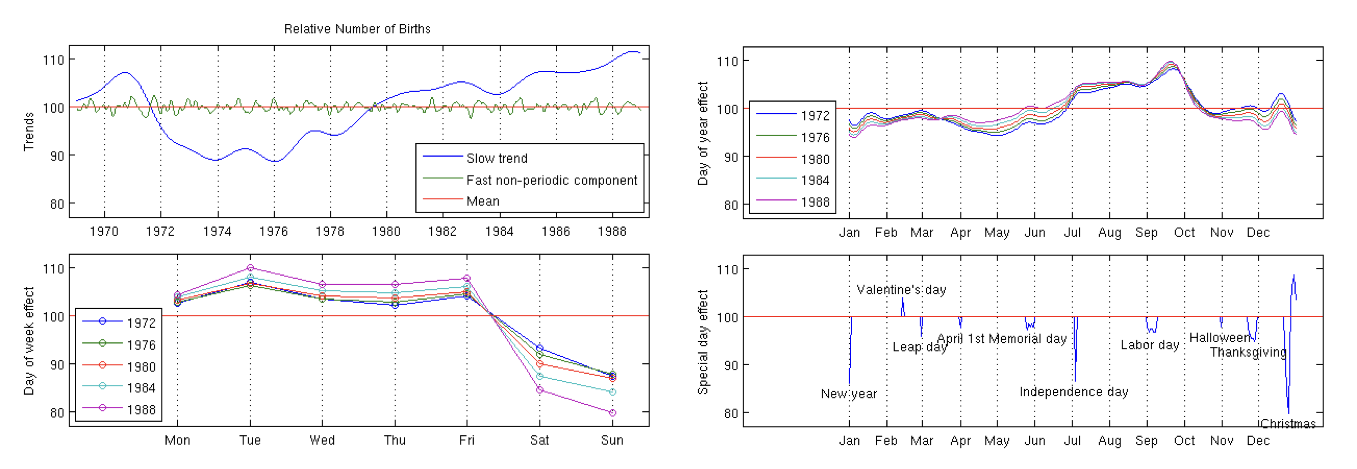
Full stan case study here with code here
slowly changing trend - yearly (sq exp cov)
small time scale trend - monthly (sq exp cov)
7 day periodic - day of week effect (periodic \(\times\) sq exp cov)
365.25 day periodic - day of year effect (periodic \(\times\) sq exp cov)
special days and interaction with weekends (linear cov)
independent Gaussian noise (nugget cov)
constant mean
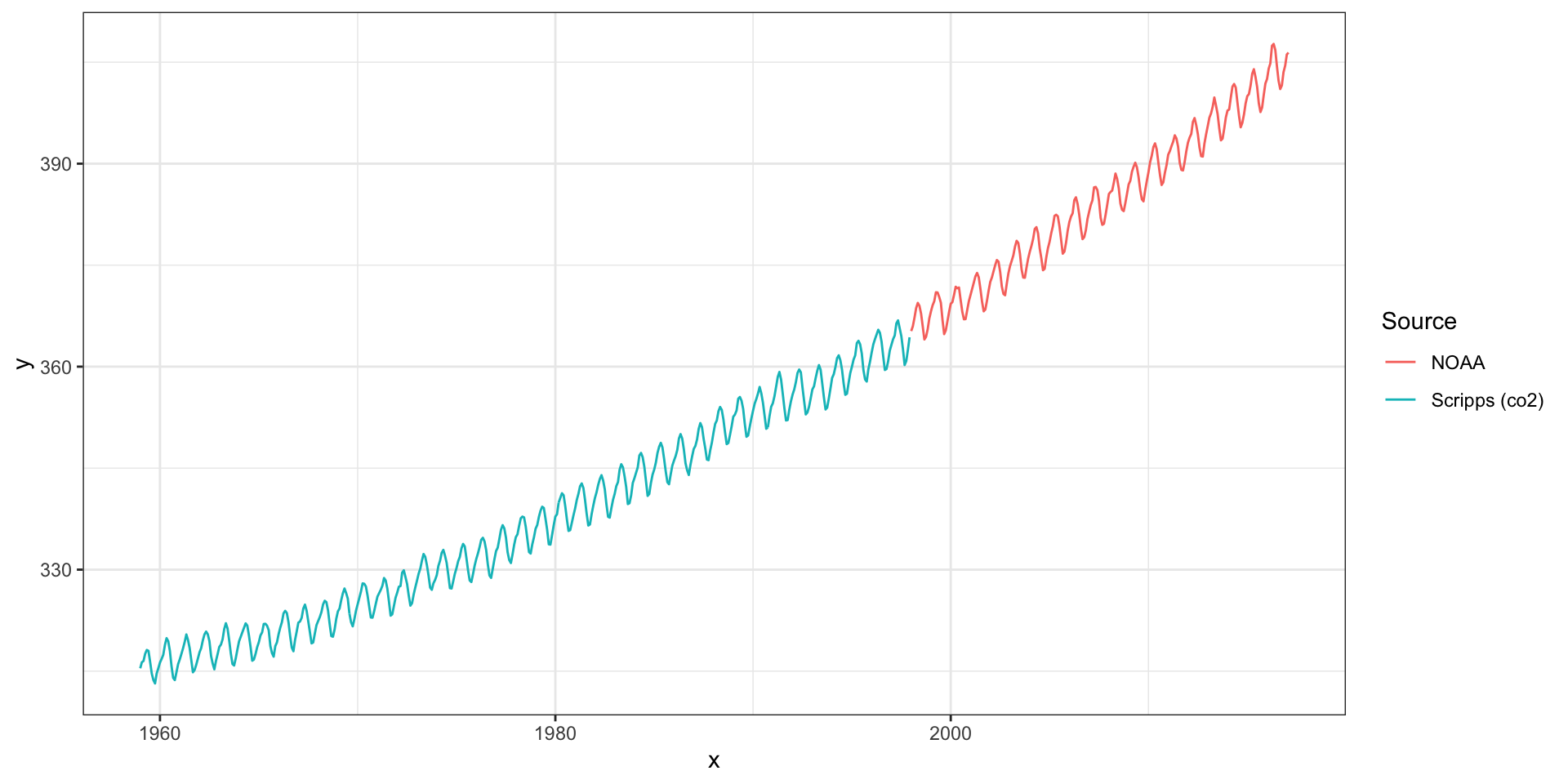
Mauna Loa Example
Atmospheric CO\(_2\)
GP Model
Based on Rasmussen 5.4.3 (we are using slightly different data and parameterization)
\[ \boldsymbol{y} \sim \mathcal{N}(\boldsymbol{\mu},~ \boldsymbol{\Sigma}_1 + \boldsymbol{\Sigma}_2 + \boldsymbol{\Sigma}_3 + \boldsymbol{\Sigma}_4 + \sigma^2 \mathit{I}\,) \]
\[\{\boldsymbol{\mu}\}_i = \bar{y}\]
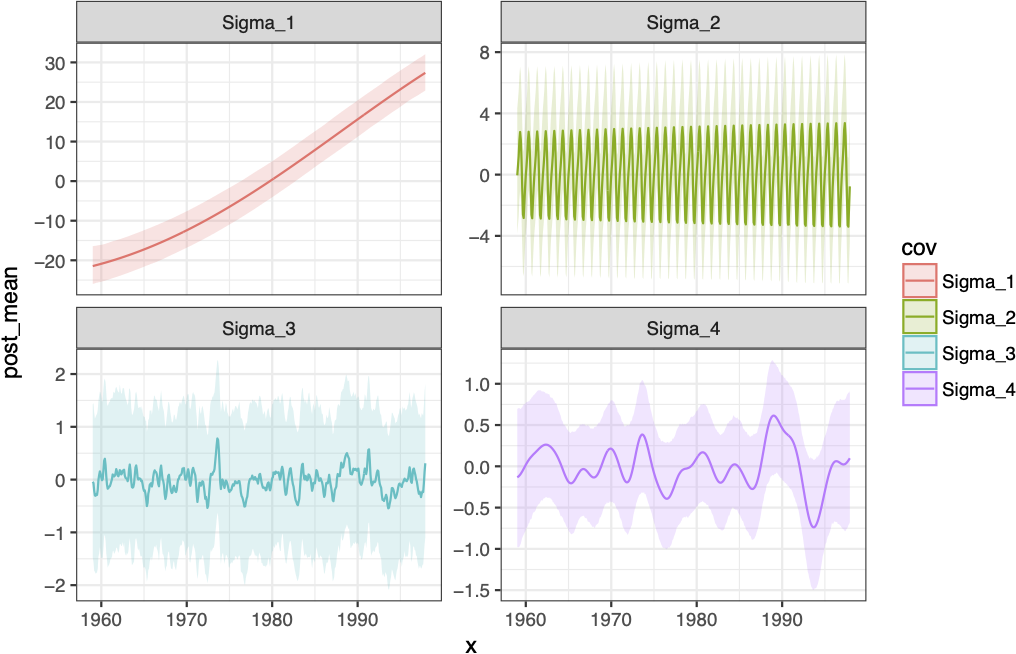
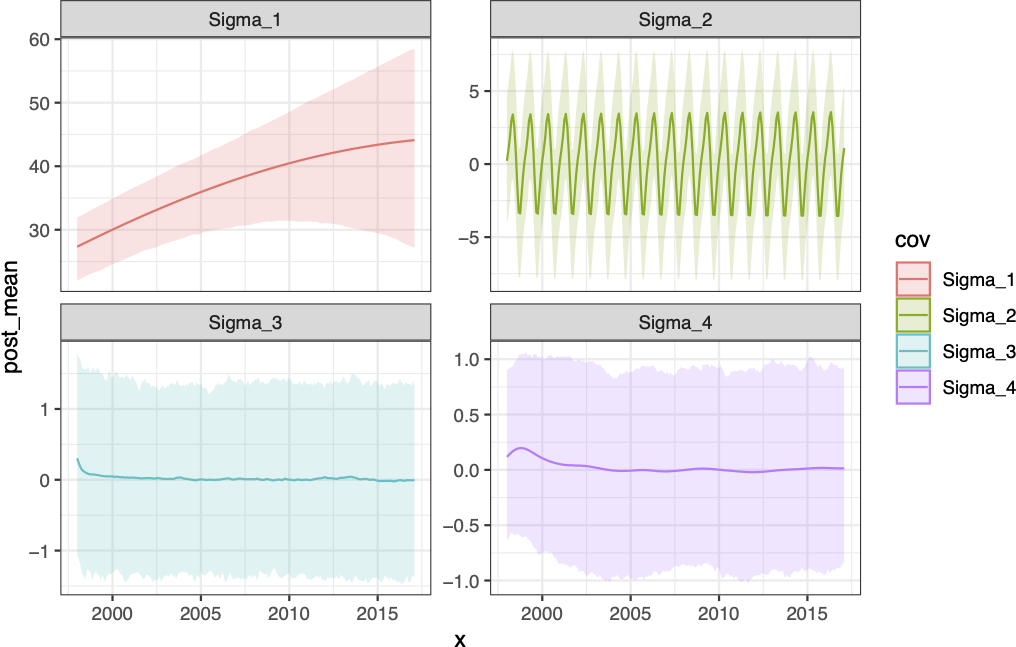
\[ \begin{aligned} \{\boldsymbol{\Sigma}_1\}_{ij} &= \sigma^2_1 \exp\left(-(l_1 \cdot d_{ij})^2\right) \\ \{\boldsymbol{\Sigma}_2\}_{ij} &= \sigma^2_2 \exp\left(-(l_2 \cdot d_{ij})^2\right)\exp\left(-2 \, (l_3)^2 \sin^2(\pi \, d_{ij} / p)\right) \\ \{\boldsymbol{\Sigma}_3\}_{ij} &= \sigma^2_3 \left(1+\frac{(l_4 \cdot d_{ij})^2}{\alpha}\right)^{-\alpha} \\ \{\boldsymbol{\Sigma}_4\}_{ij} &= \sigma^2_4 \exp\left(-(l_5 \cdot d_{ij})^2\right) \end{aligned} \]
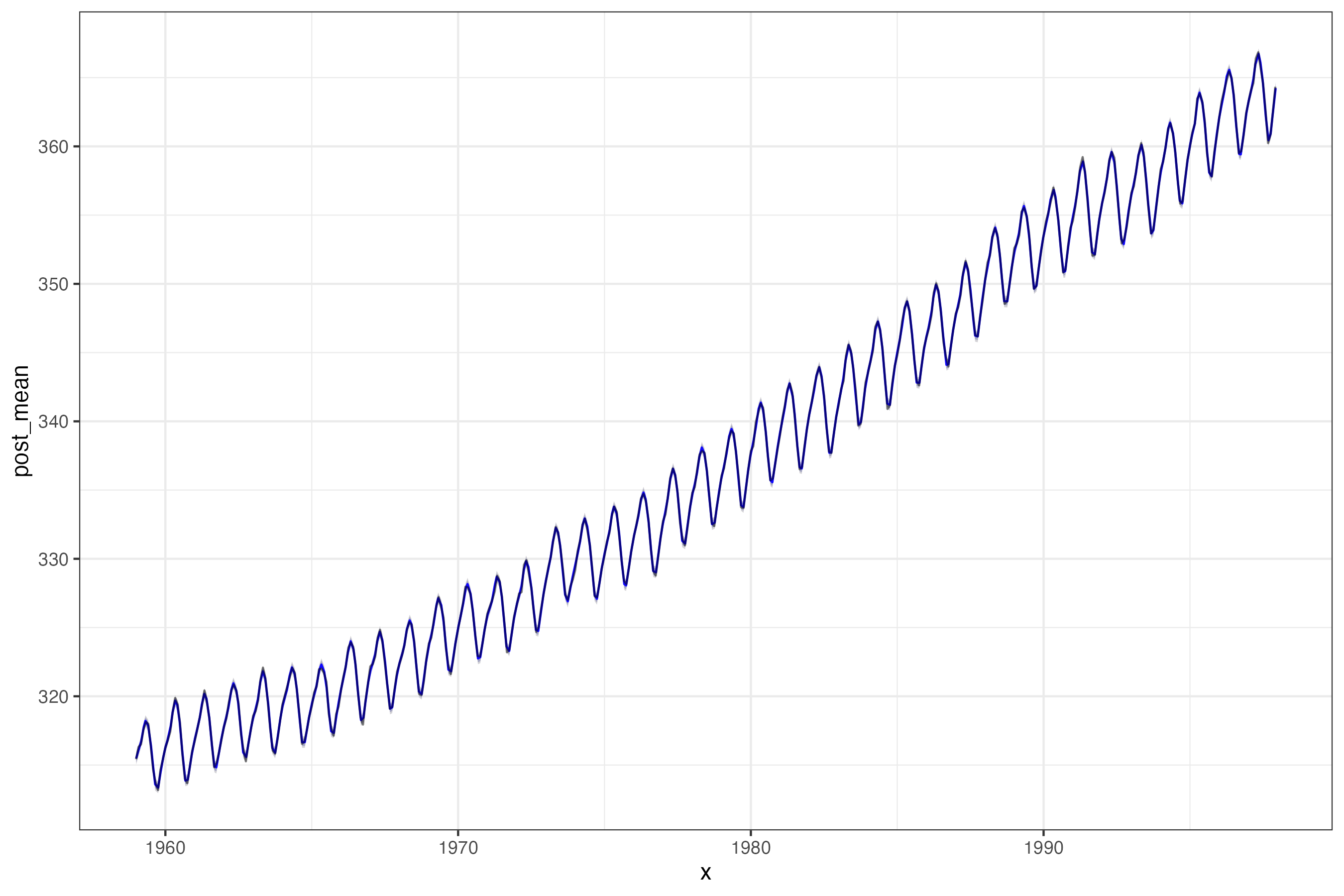
Model fit
Fit Components
Model fit + forecast
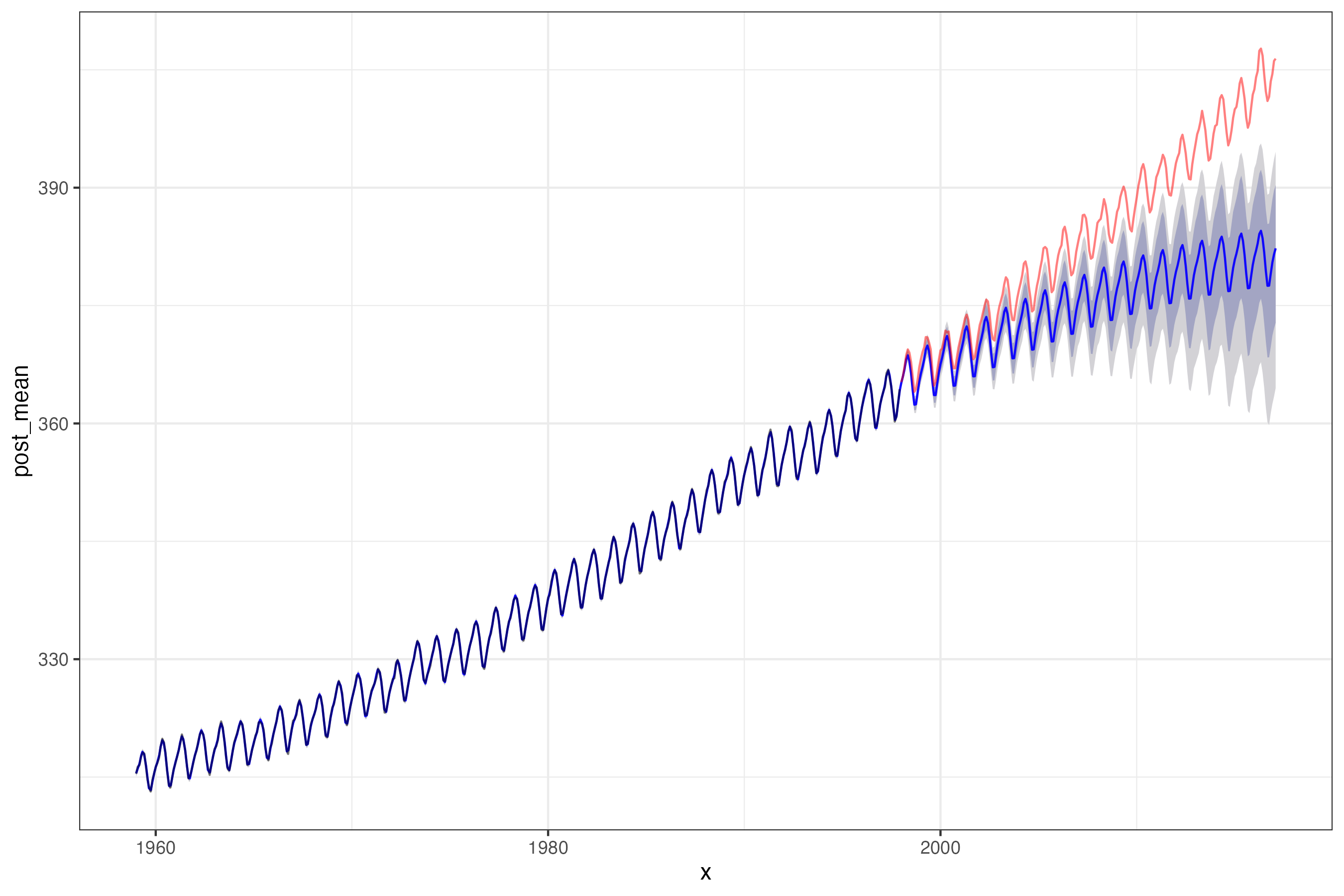
Forecast
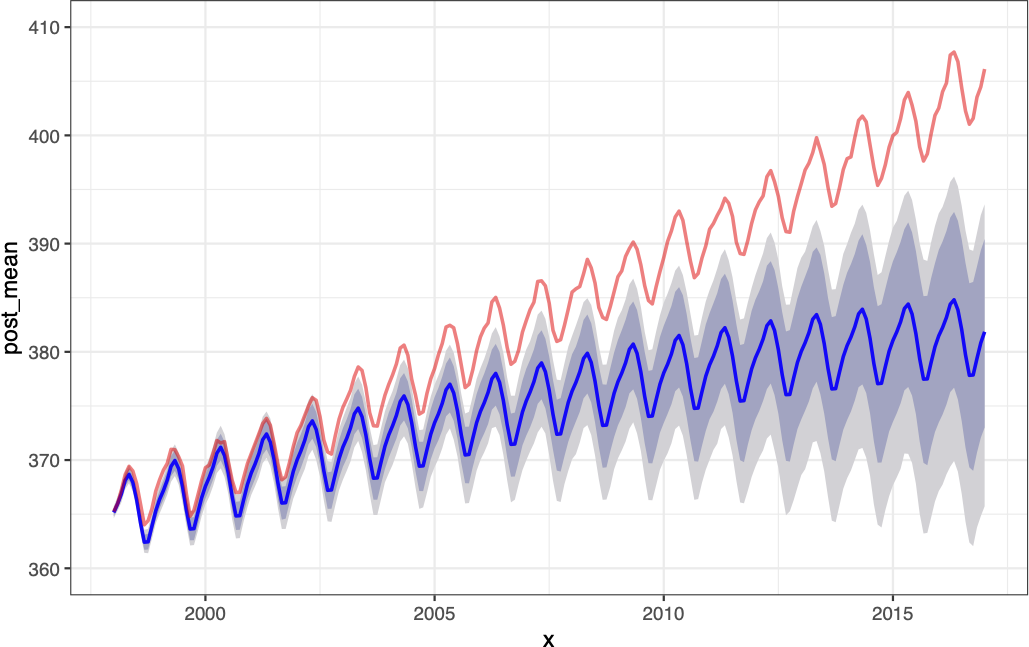
Forecast components
ARIMA forecast
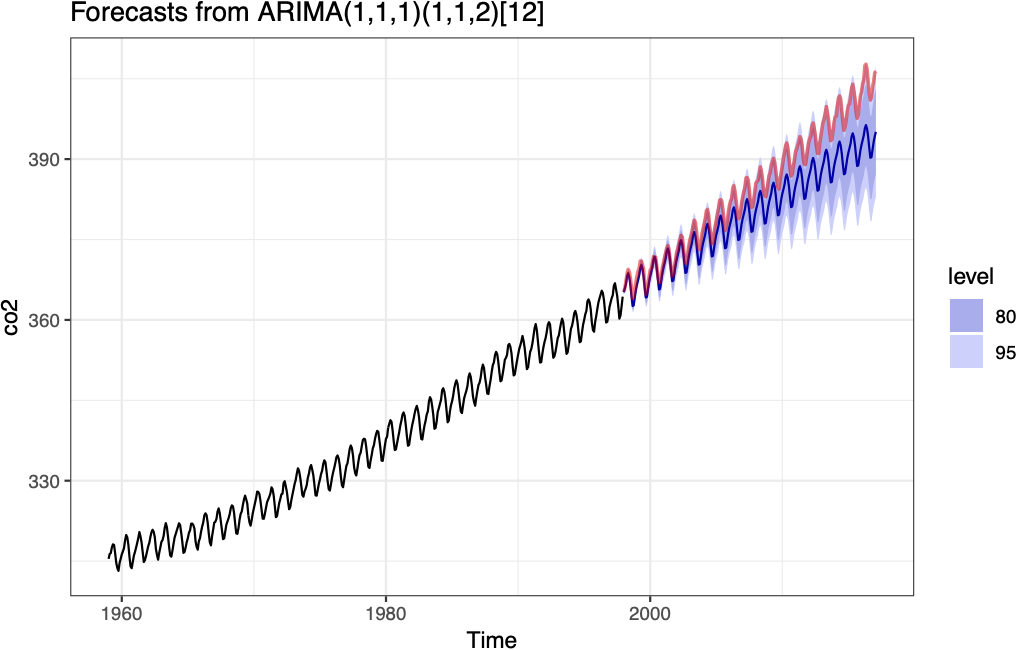
Model performance
| Forecast dates |
arima RMSE |
gp RMSE |
|---|---|---|
| Jan 1998 - Jan 2003 | 1.10 | 1.91 |
| Jan 1998 - Jan 2008 | 2.51 | 4.58 |
| Jan 1998 - Jan 2013 | 3.82 | 7.71 |
| Jan 1998 - Mar 2017 | 5.46 | 11.40 |
Sta 344 - Fall 2022