Gaussian Process Models
Lecture 14
Multivariate Normal
Multivariate Normal Distribution
For an \(n\)-dimension multivate normal distribution with covariance \(\boldsymbol{\Sigma}\) (positive semidefinite) can be written as
\[ \underset{n \times 1}{\boldsymbol{y}} \sim N(\underset{n \times 1}{\boldsymbol{\mu}}, \; \underset{n \times n}{\boldsymbol{\Sigma}}) \]
\[ \begin{pmatrix} y_1\\ \vdots\\ y_n \end{pmatrix} \sim N\left( \begin{pmatrix} \mu_1\\ \vdots\\ \mu_n \end{pmatrix}, \, \begin{pmatrix} \rho_{11}\sigma_1\sigma_1 & \cdots & \rho_{1n}\sigma_1\sigma_n \\ \vdots & \ddots & \vdots \\ \rho_{n1}\sigma_n\sigma_1 & \cdots & \rho_{nn}\sigma_n\sigma_n \\ \end{pmatrix} \right) \]
Density
For the \(n\) dimensional multivate normal given on the last slide, its density is given by
\[ (2\pi)^{-n/2} \; \det(\boldsymbol{\Sigma})^{-1/2} \; \exp{\left(-\frac{1}{2} \underset{1 \times n}{(\boldsymbol{y}-\boldsymbol{\mu})'} \underset{n \times n}{\boldsymbol{\Sigma}^{-1}} \underset{n \times 1}{(\boldsymbol{y}-\boldsymbol{\mu})}\right)} \]
and its log density is given by
\[ -\frac{n}{2} \log 2\pi - \frac{1}{2} \log \det(\boldsymbol{\Sigma}) - -\frac{1}{2} \underset{1 \times n}{(\boldsymbol{y}-\boldsymbol{\mu})'} \underset{n \times n}{\boldsymbol{\Sigma}^{-1}} \underset{n \times 1}{(\boldsymbol{y}-\boldsymbol{\mu})} \]
Sampling
To generate draws from an \(n\)-dimensional multivate normal with mean \(\underset{n \times 1}{\boldsymbol{\mu}}\) and covariance matrix \(\underset{n \times n}{\boldsymbol{\Sigma}}\),
Find a matrix \(\underset{n \times n}{\boldsymbol{A}}\) such that \(\boldsymbol{\Sigma} = \boldsymbol{A}\,\boldsymbol{A}^t\)
- most often we use \(\boldsymbol{A} = \text{Chol}(\boldsymbol{\Sigma})\) where \(\boldsymbol{A}\) is a lower triangular matrix.
- Draw \(n\) iid unit normals, \(N(0,1)\), as \(\underset{n \times 1}{\boldsymbol{z}}\)
- Obtain multivariate normal draws using \[ \underset{n \times 1}{\boldsymbol{y}} = \underset{n \times 1}{\boldsymbol{\mu}} + \underset{n \times n}{\boldsymbol{A}} \, \underset{n \times 1}{\boldsymbol{z}} \]
Bivariate Examples
\[ \boldsymbol{\mu} = \begin{pmatrix}0 \\ 0\end{pmatrix} \qquad \boldsymbol{\Sigma} = \begin{pmatrix}1 & \rho \\ \rho & 1 \end{pmatrix}\]
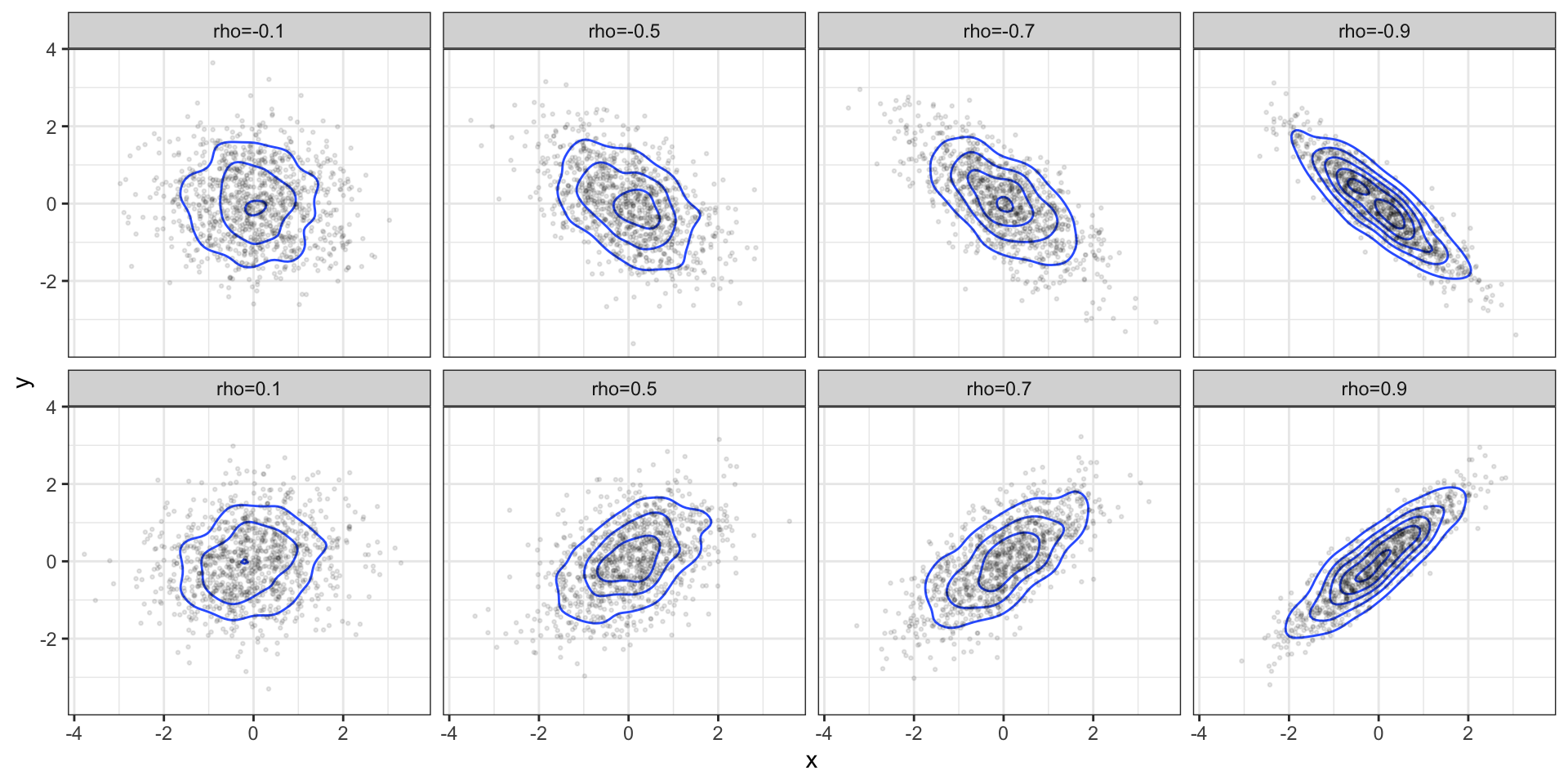
Marginal / conditional distributions
Proposition - For an \(n\)-dimensional multivate normal with mean \(\boldsymbol{\mu}\) and covariance matrix \(\boldsymbol{\Sigma}\), any marginal or conditional distribution of the \(y\)’s will also be (multivariate) normal.
Univariate marginal distribution: \[ y_i = N(\boldsymbol{\mu}_i,\,\boldsymbol{\Sigma}_{ii}) \]
Bivariate marginal distribution: \[ \boldsymbol{y}_{ij} = N\left( \begin{pmatrix}\boldsymbol{\mu}_i \\ \boldsymbol{\mu}_j \end{pmatrix},\; \begin{pmatrix} \boldsymbol{\Sigma}_{ii} & \boldsymbol{\Sigma}_{ij} \\ \boldsymbol{\Sigma}_{ji} & \boldsymbol{\Sigma}_{jj} \end{pmatrix} \right) \]
\(k\)-dimensional marginal distribution:
\[ \boldsymbol{y}_{i,\ldots,k} = N\left( \begin{pmatrix}\boldsymbol{\mu}_{i} \\ \vdots \\ \boldsymbol{\mu}_{k} \end{pmatrix},\; \begin{pmatrix} \boldsymbol{\Sigma}_{ii} & \cdots & \boldsymbol{\Sigma}_{i k} \\ \vdots & \ddots & \vdots \\ \boldsymbol{\Sigma}_{k i} & \cdots & \boldsymbol{\Sigma}_{k k} \end{pmatrix} \right) \]
Conditional Distributions
If we partition the \(n\)-dimensions into two pieces such that \(\boldsymbol{y} = (\boldsymbol{y}_1,\, \boldsymbol{y}_2)^t\) then
\[ \underset{n \times 1}{\boldsymbol{y}} \sim N\left( \begin{pmatrix} \boldsymbol{\mu}_1 \\ \boldsymbol{\mu}_2 \end{pmatrix},\, \begin{pmatrix} \boldsymbol{\Sigma}_{11} & \boldsymbol{\Sigma}_{12} \\ \boldsymbol{\Sigma}_{21} & \boldsymbol{\Sigma}_{22} \end{pmatrix} \right) \] \[ \begin{aligned} \underset{k \times 1}{~~\boldsymbol{y}_1~~} &\sim N(\underset{k \times 1}{~~~\boldsymbol{\mu}_1~~~},\, \underset{k \times k}{~~~\boldsymbol{\Sigma}_{11}~~~}) \\ \underset{n-k \times 1}{\boldsymbol{y}_2} &\sim N(\underset{n-k \times 1}{\boldsymbol{\mu}_2},\, \underset{n-k \times n-k}{\boldsymbol{\Sigma}_{22}}) \end{aligned} \]
then the conditional distributions are given by
\[ \begin{aligned} \boldsymbol{y}_1 ~|~ \boldsymbol{y}_2 = \boldsymbol{a} ~&\sim N(\boldsymbol{\mu_1} + \boldsymbol{\Sigma_{12}} \, \boldsymbol{\Sigma_{22}}^{-1} \, (\boldsymbol{a} - \boldsymbol{\mu_2}),~ \boldsymbol{\Sigma_{11}}-\boldsymbol{\Sigma_{12}}\,\boldsymbol{\Sigma_{22}}^{-1} \, \boldsymbol{\Sigma_{21}}) \\ \\ \boldsymbol{y}_2 ~|~ \boldsymbol{y}_1 = \boldsymbol{b} ~&\sim N(\boldsymbol{\mu_2} + \boldsymbol{\Sigma_{21}} \, \boldsymbol{\Sigma_{11}}^{-1} \, (\boldsymbol{b} - \boldsymbol{\mu_1}),~ \boldsymbol{\Sigma_{22}}-\boldsymbol{\Sigma_{21}}\,\boldsymbol{\Sigma_{11}}^{-1} \, \boldsymbol{\Sigma_{12}}) \end{aligned} \]
Gaussian Processes
From Shumway,
A process, \(\boldsymbol{y} = \{y(t) ~:~ t \in T\}\), is said to be a Gaussian process if all possible finite dimensional vectors \(\boldsymbol{y} = (y_{t_1},y_{t_2},...,y_{t_n})^t\), for every collection of time points \(t_1, t_2, \ldots , t_n\), and every positive integer \(n\), have a multivariate normal distribution.
So far we have only looked at examples of time series where \(T\) is discete (and evenly spaces & contiguous), it turns out things get a lot more interesting when we explore the case where \(T\) is defined on a continuous space (e.g. \(\mathbb{R}\) or some subset of \(\mathbb{R}\)).
Gaussian Process Regression
Parameterizing a Gaussian Process
Imagine we have a Gaussian process defined such that \(\boldsymbol{y} = \{y(t) ~:~ t \in [0,1]\}\),
- We now have an uncountably infinite set of possible \(t\)’s and \(y(t)\)s.
- We will only have a (small) finite number of observations \(y(t_1), \ldots, y(t_n)\) with which to say something useful about this infinite dimensional process.
- The unconstrained covariance matrix for the observed data can have up to \(n(n+1)/2\) unique values\(^*\)
Necessary to make some simplifying assumptions:
Stationarity
Simple(r) parameterization of \(\Sigma\)
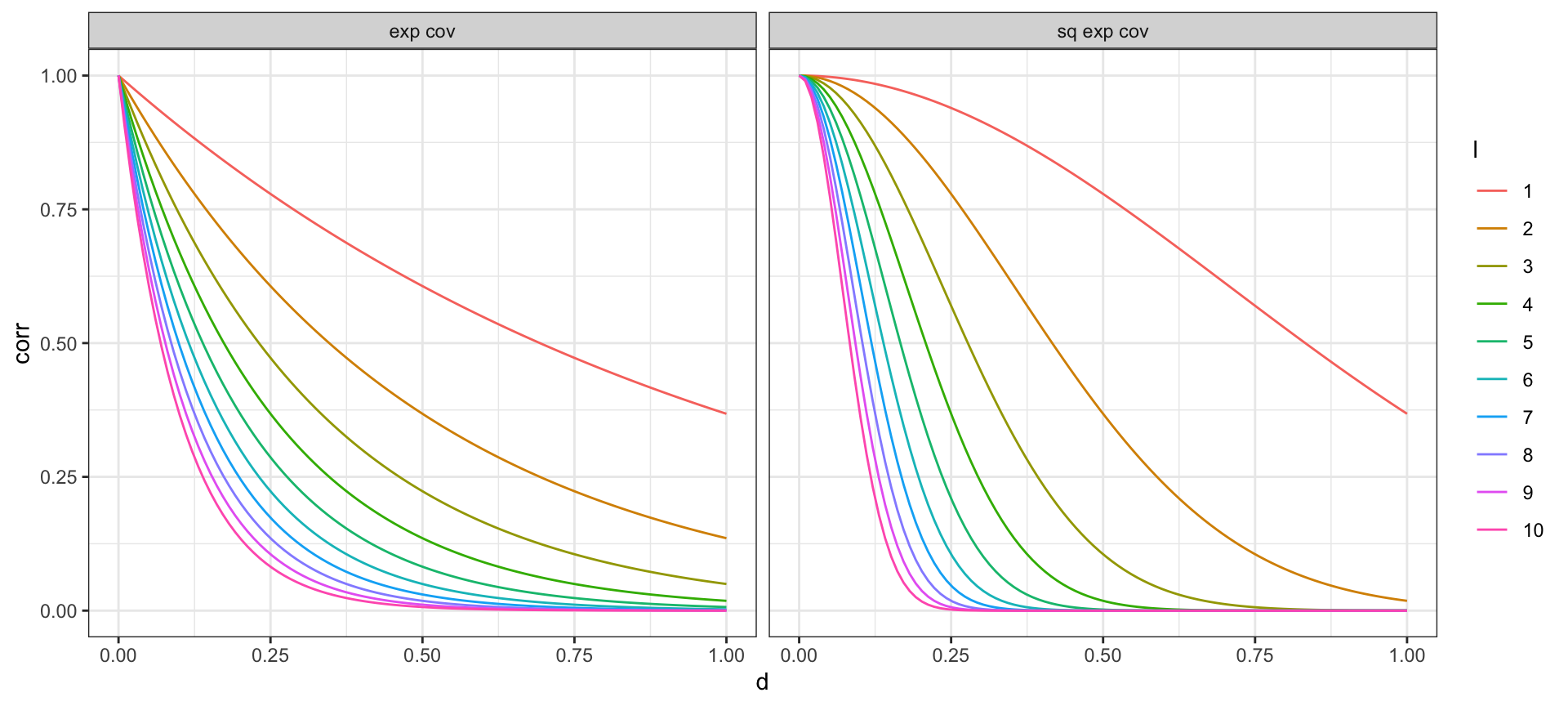
Covariance Functions
More on these next week, but for now some common examples
Exponential covariance: \[ \Sigma(y_{t},y_{t'}) = \sigma^2 \exp\big(-|t-t'| \; l\,\big) \]
Squared exponential covariance (Gaussian): \[ \Sigma(y_{t},y_{t'}) = \sigma^2 \exp\big(-(|t-t'| \; l\,)^2\big) \]
Powered exponential covariance \(\left(p \in (0,2]\right)\): \[ \Sigma(y_{t},y_{t'}) = \sigma^2 \exp\big(-(|t-t'| \; l\,)^p\big) \]
Covariance Function - Correlation Decay
Letting \(\sigma^2 = 1\) and trying different values of the length scale \(l\),
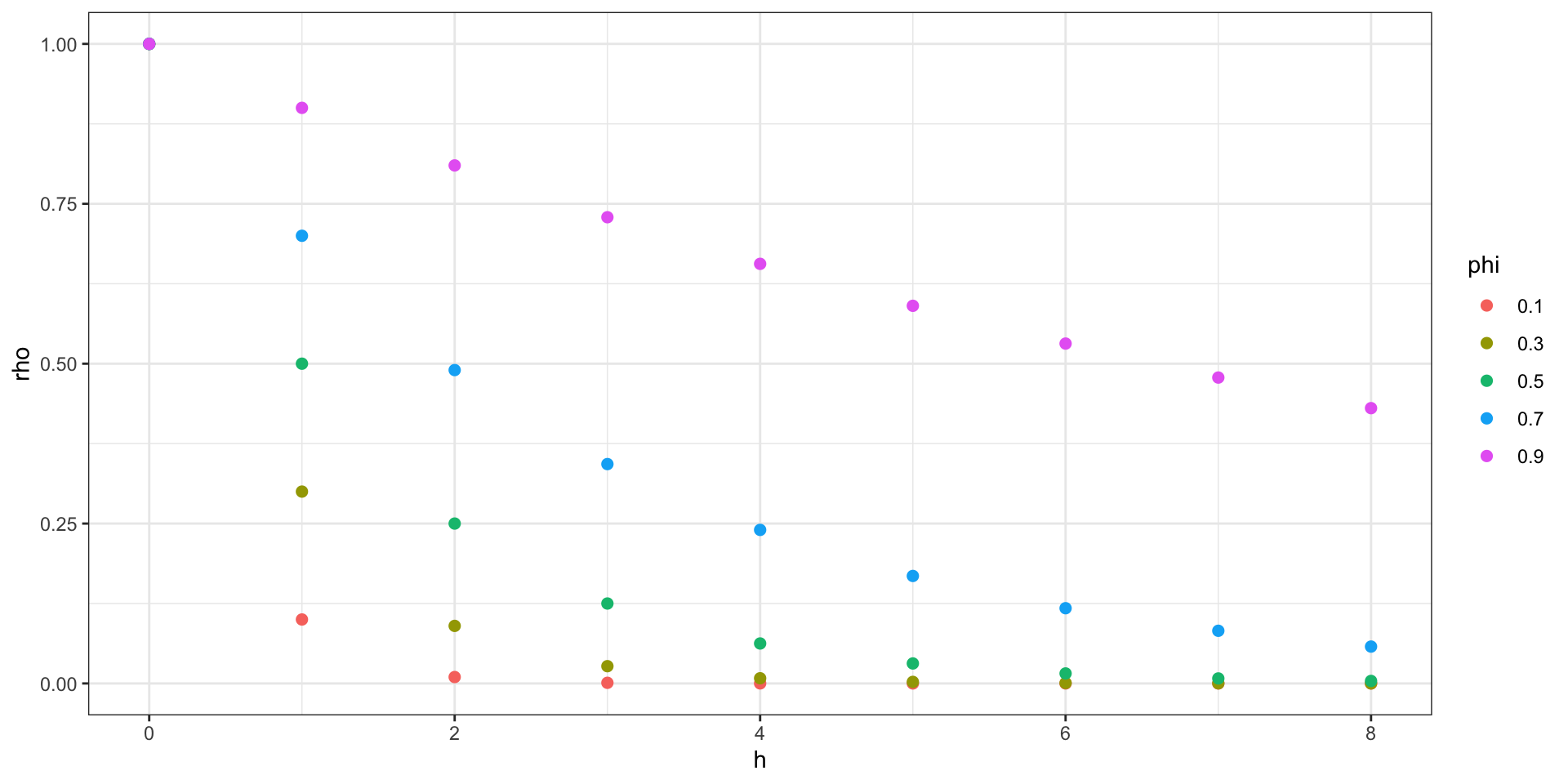
Correlation Decay - AR(1)
Recall that for a stationary AR(1) process: \(\gamma(h) = \sigma^2_w \phi^{|h|}\) and \(\rho(h) = \phi^{|h|}\)
we can draw a somewhat similar picture about the decay of \(\rho\) as a function of distance.
Example
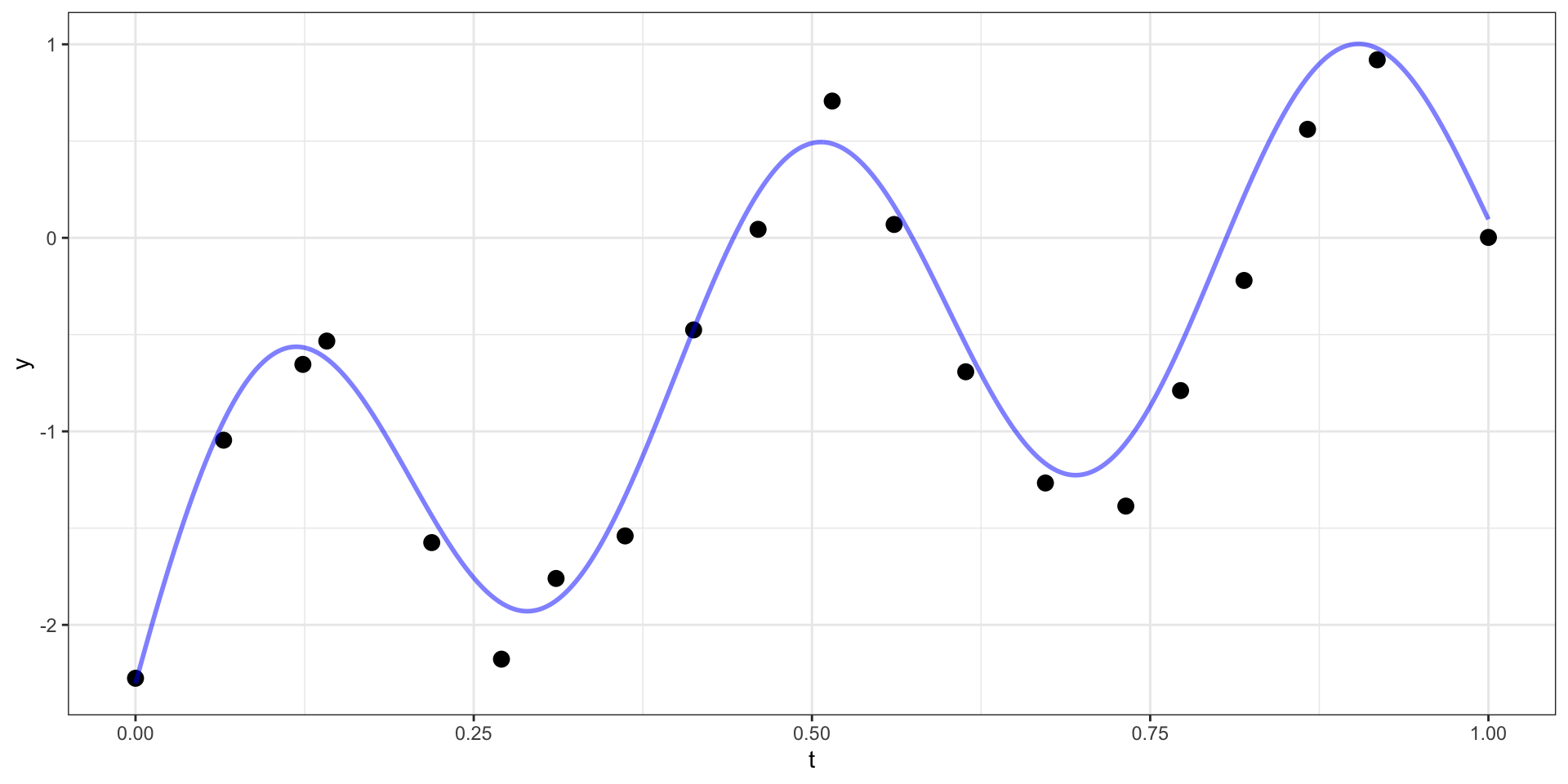
Prediction
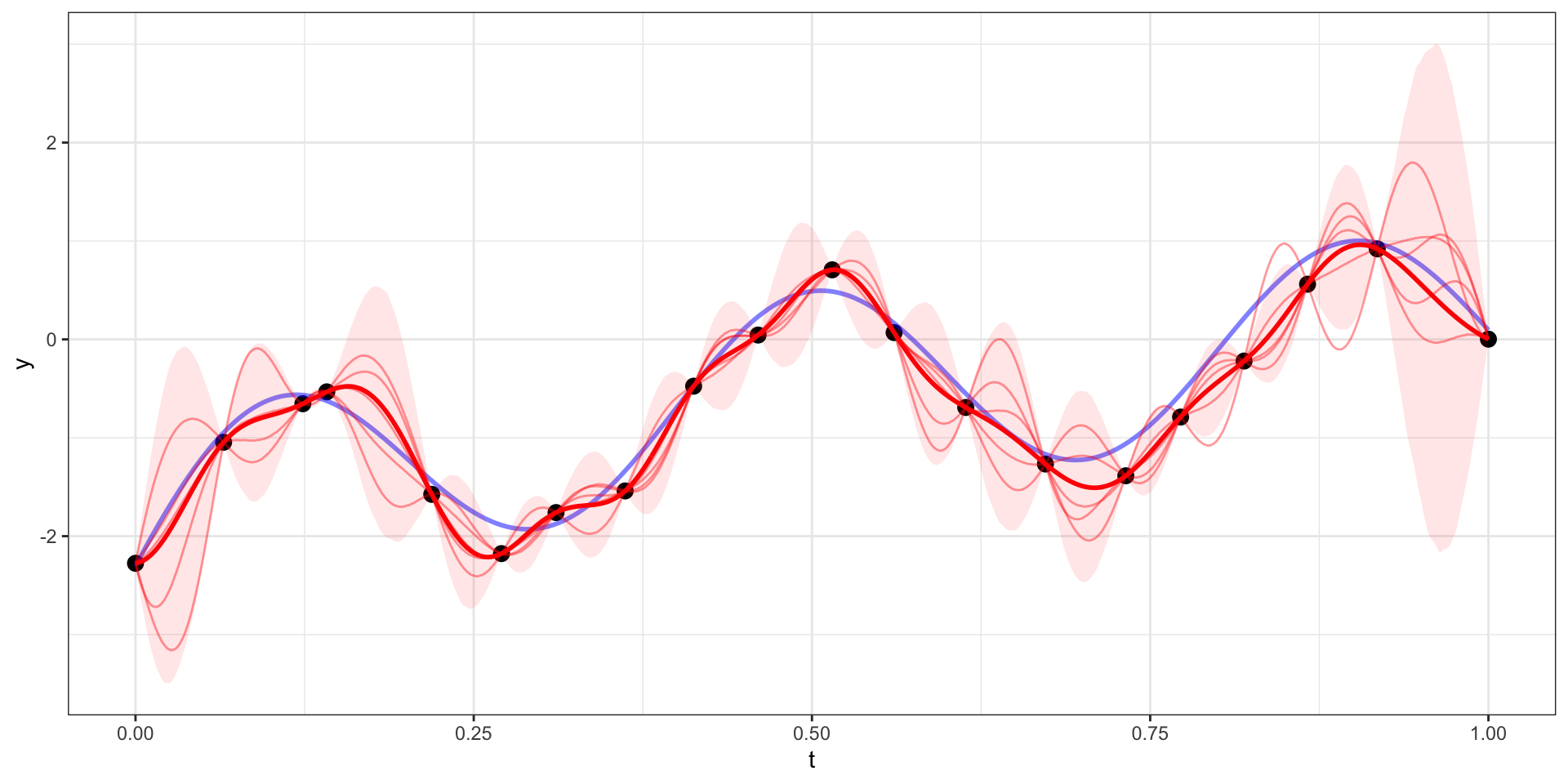
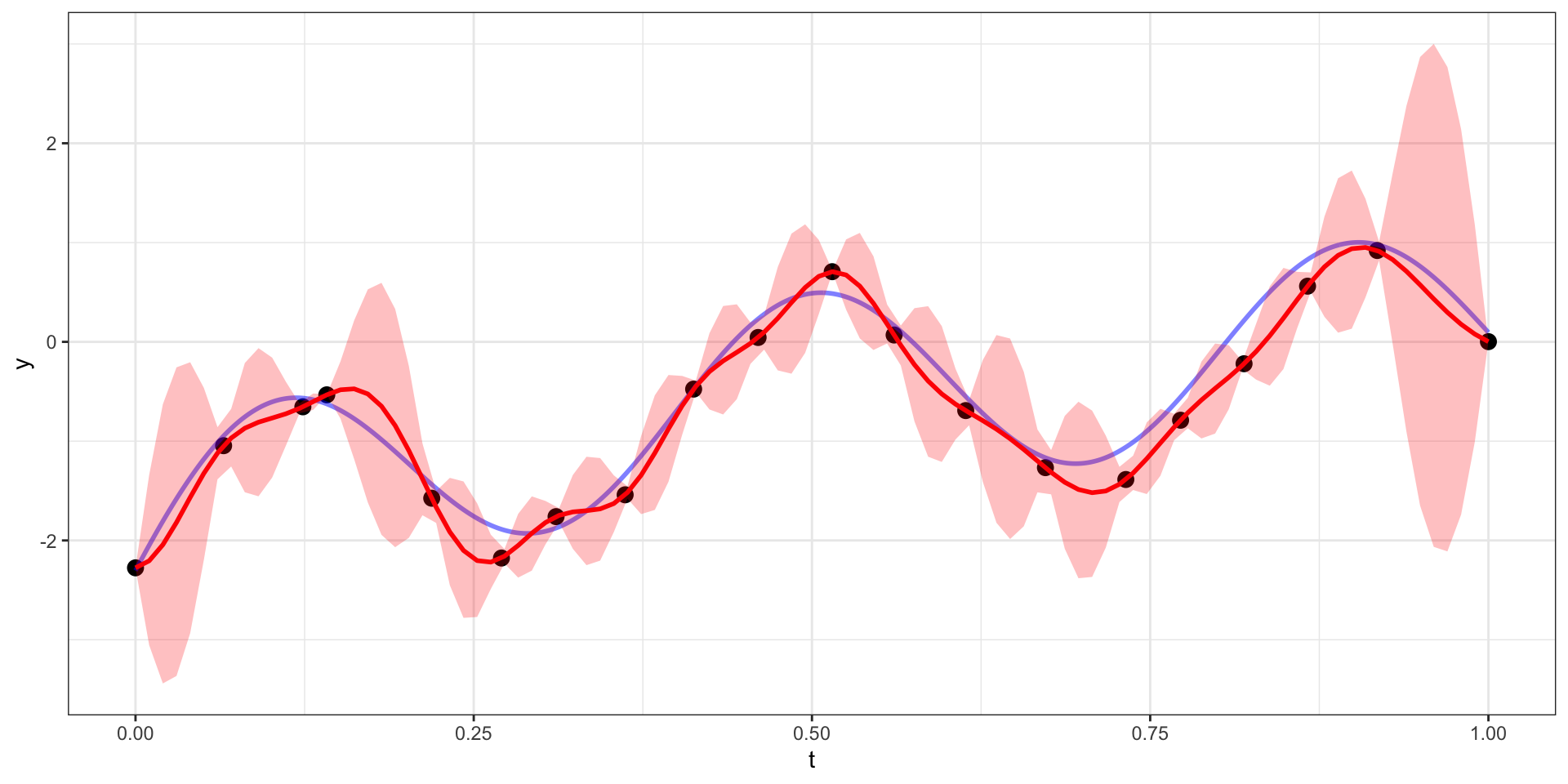
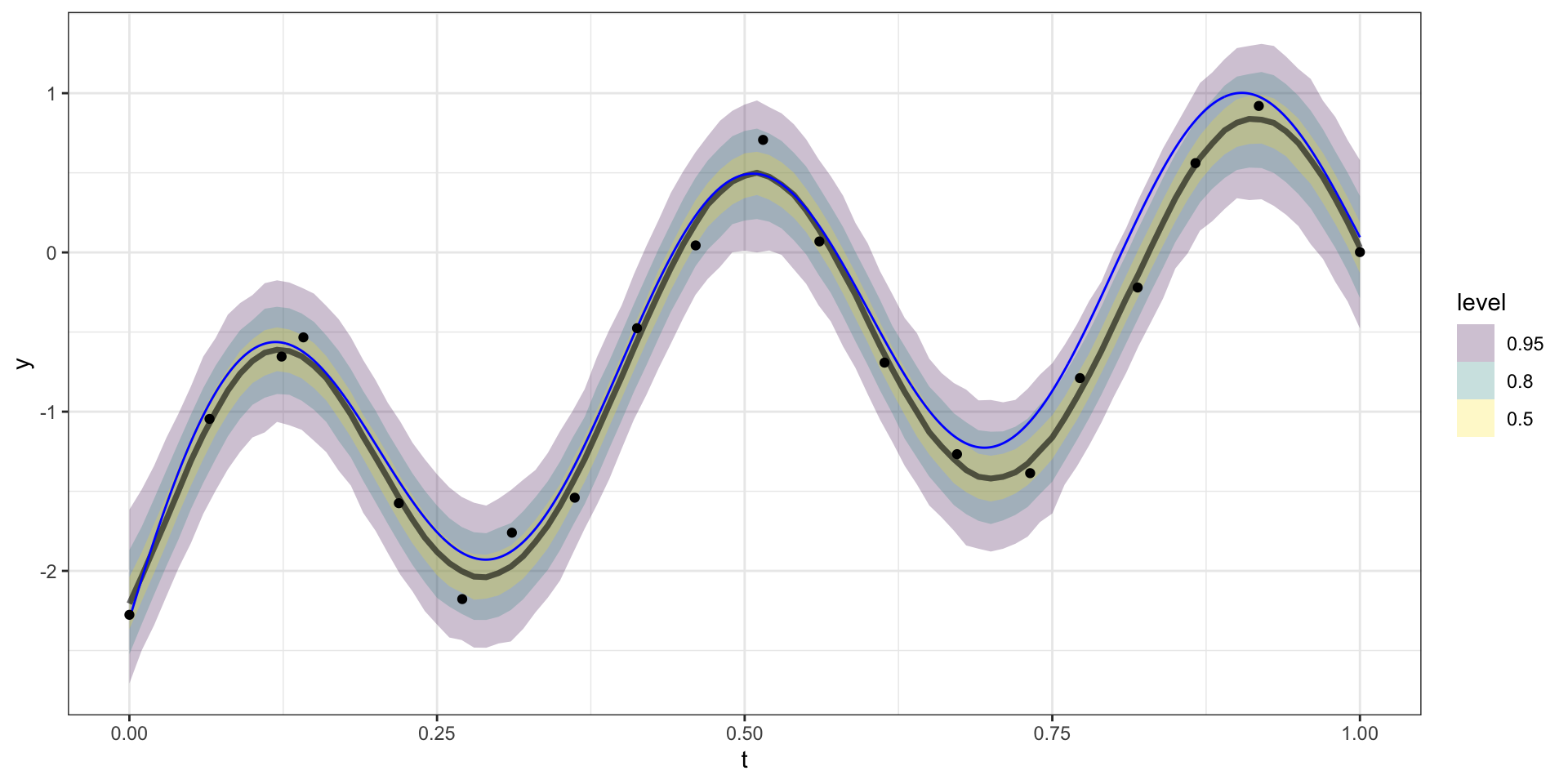
Our example has 20 observations which we would like to use as the basis for predicting \(y(t)\) at other values of \(t\) (say a regular sequence of values from 0 to 1).
For now lets use a square exponential covariance with \(\sigma^2 = 10\) and \(l=15\)
We therefore want to sample from \(\boldsymbol{y}_{pred} | \boldsymbol{y}_{obs}\).
\[\boldsymbol{y}_{pred} ~|~ \boldsymbol{y}_{obs} = \boldsymbol{y} ~\sim N(\boldsymbol{\Sigma}_{po} \, \boldsymbol{\Sigma}_{obs}^{-1} \, \boldsymbol{y},~ \boldsymbol{\Sigma}_{pred}-\boldsymbol{\Sigma}_{po}\,\boldsymbol{\Sigma}_{pred}^{-1} \, \boldsymbol{\Sigma}_{op})\]
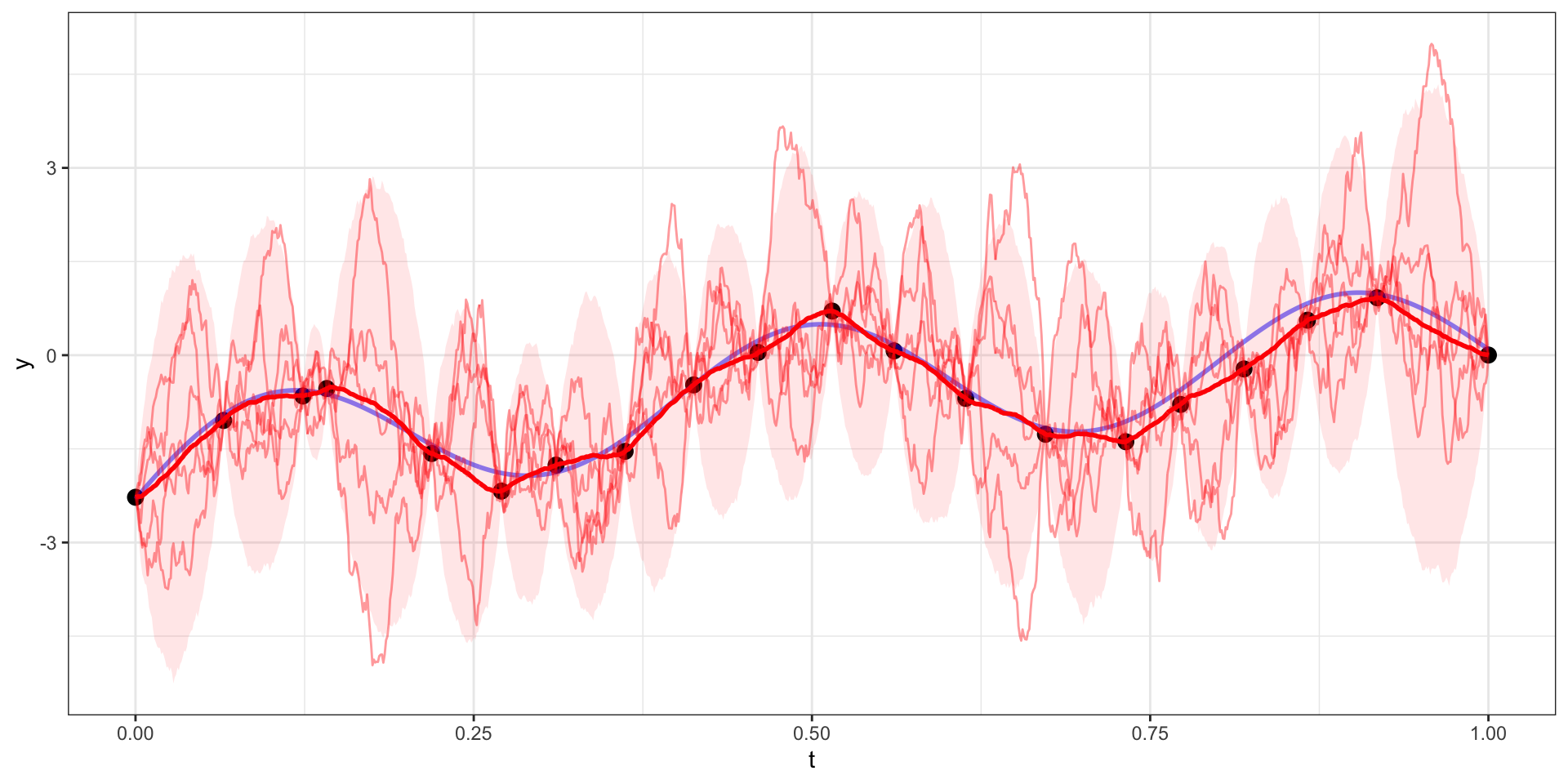
Draw 1
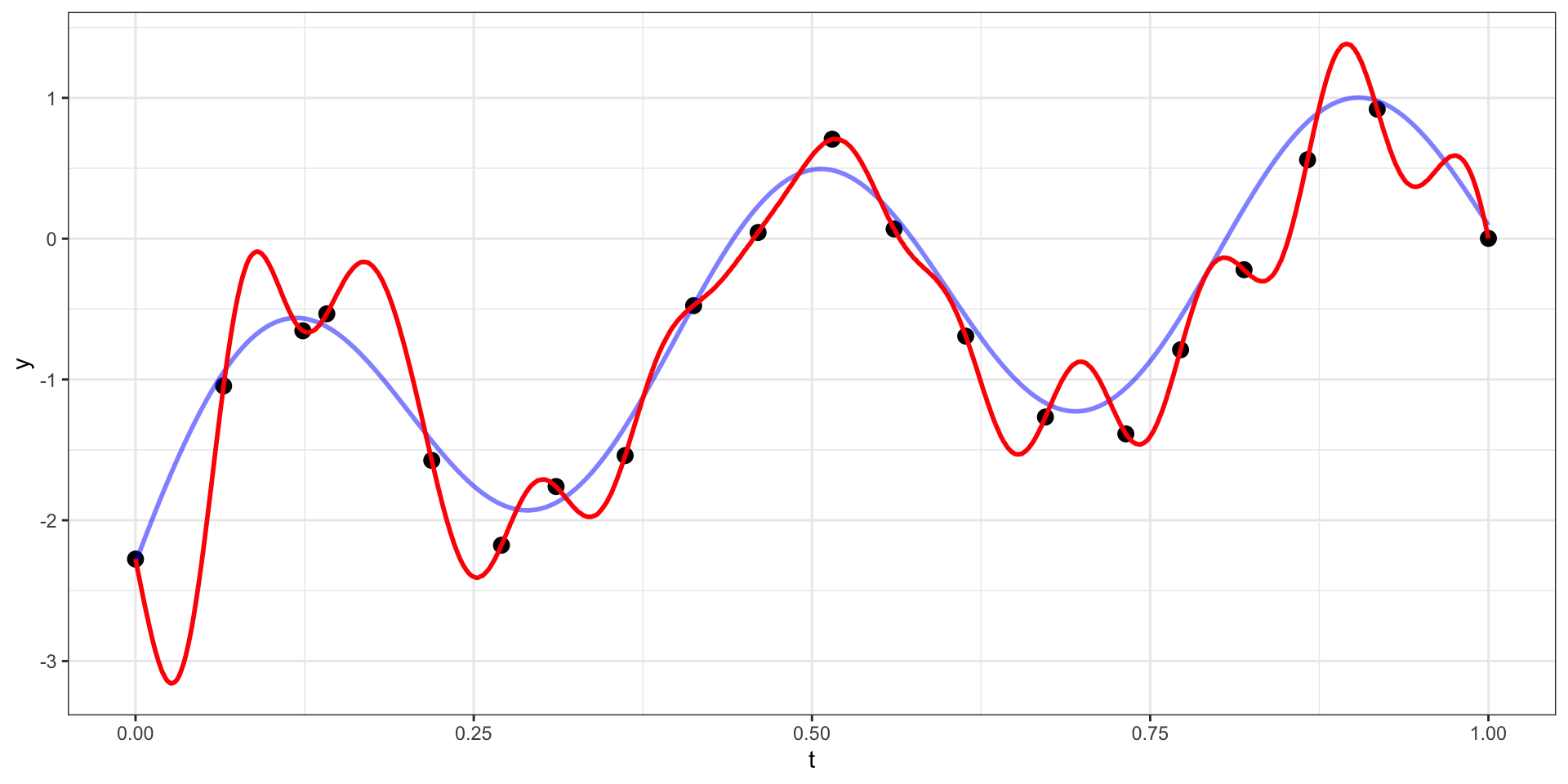
Draw 2
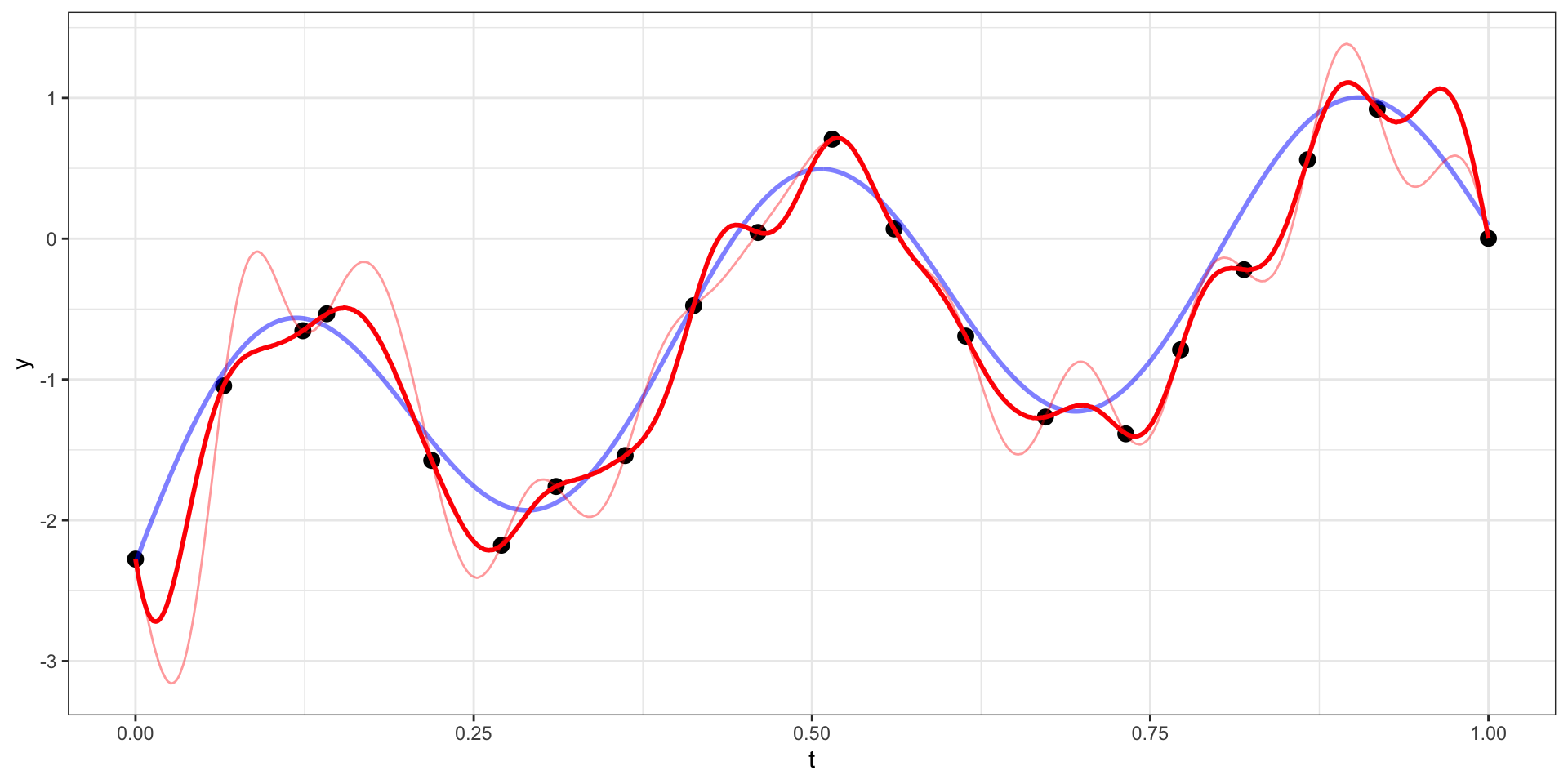
Draw 3
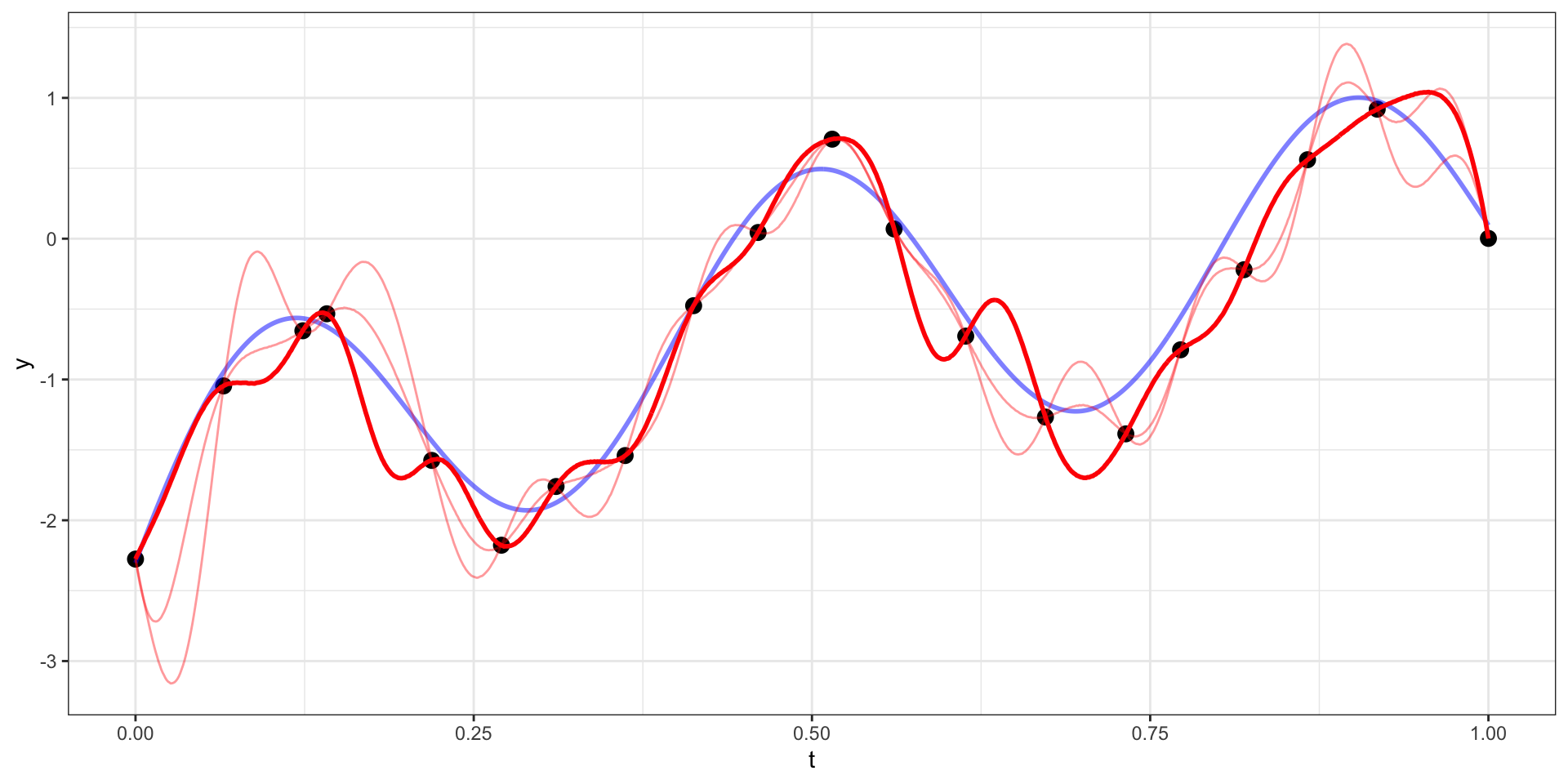
Draw 4
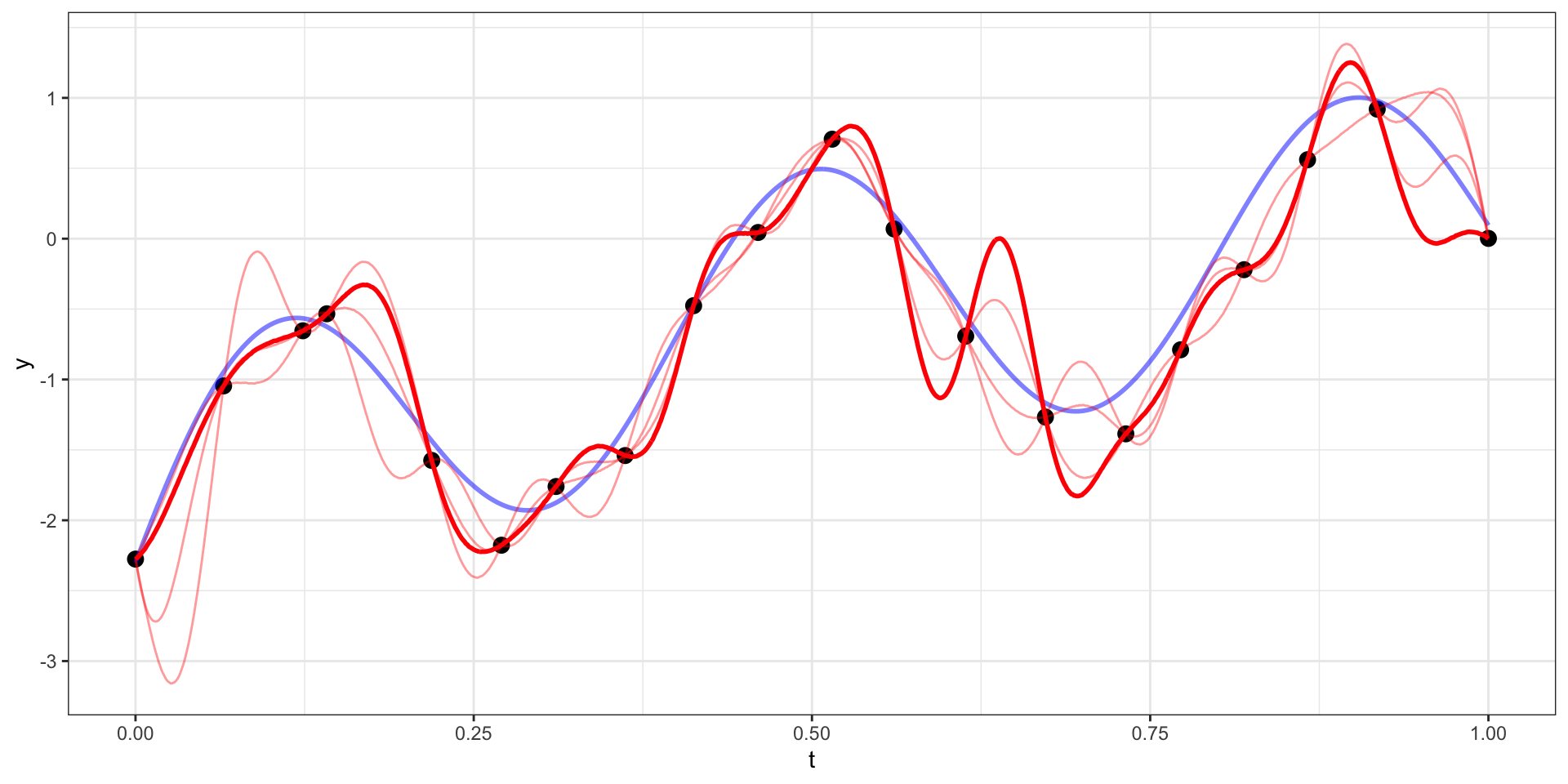
Draw 5
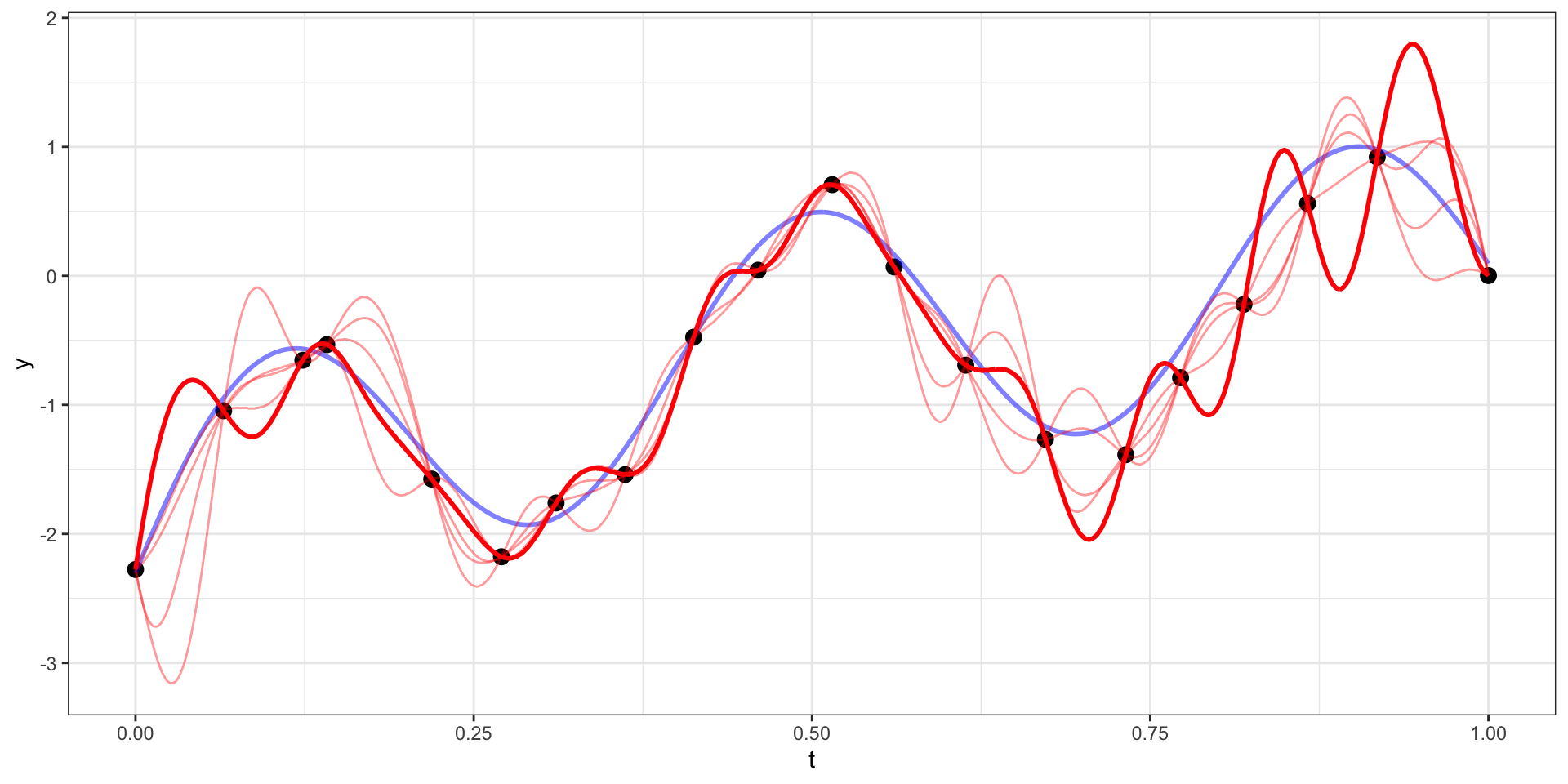
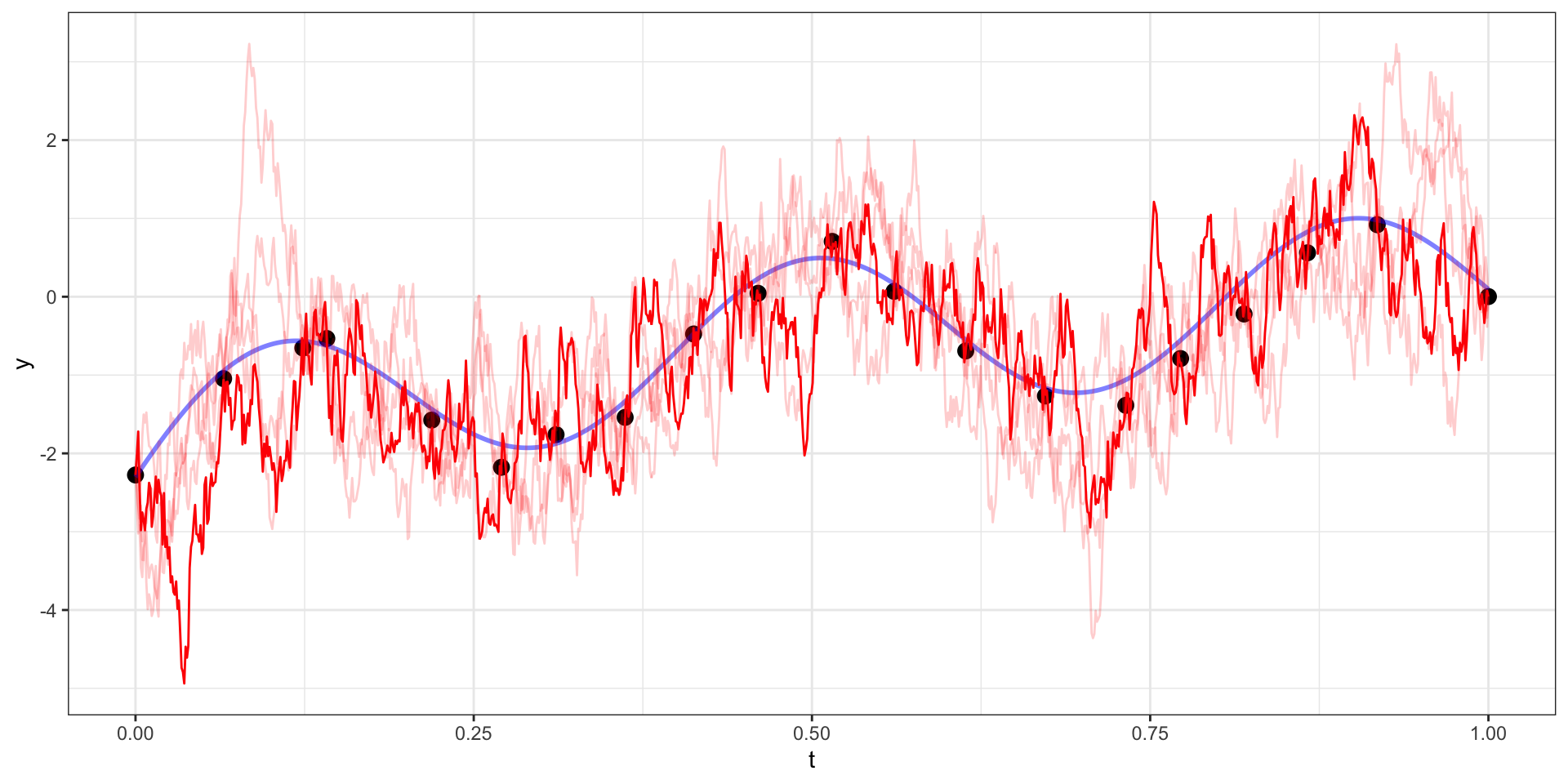
Many draws later
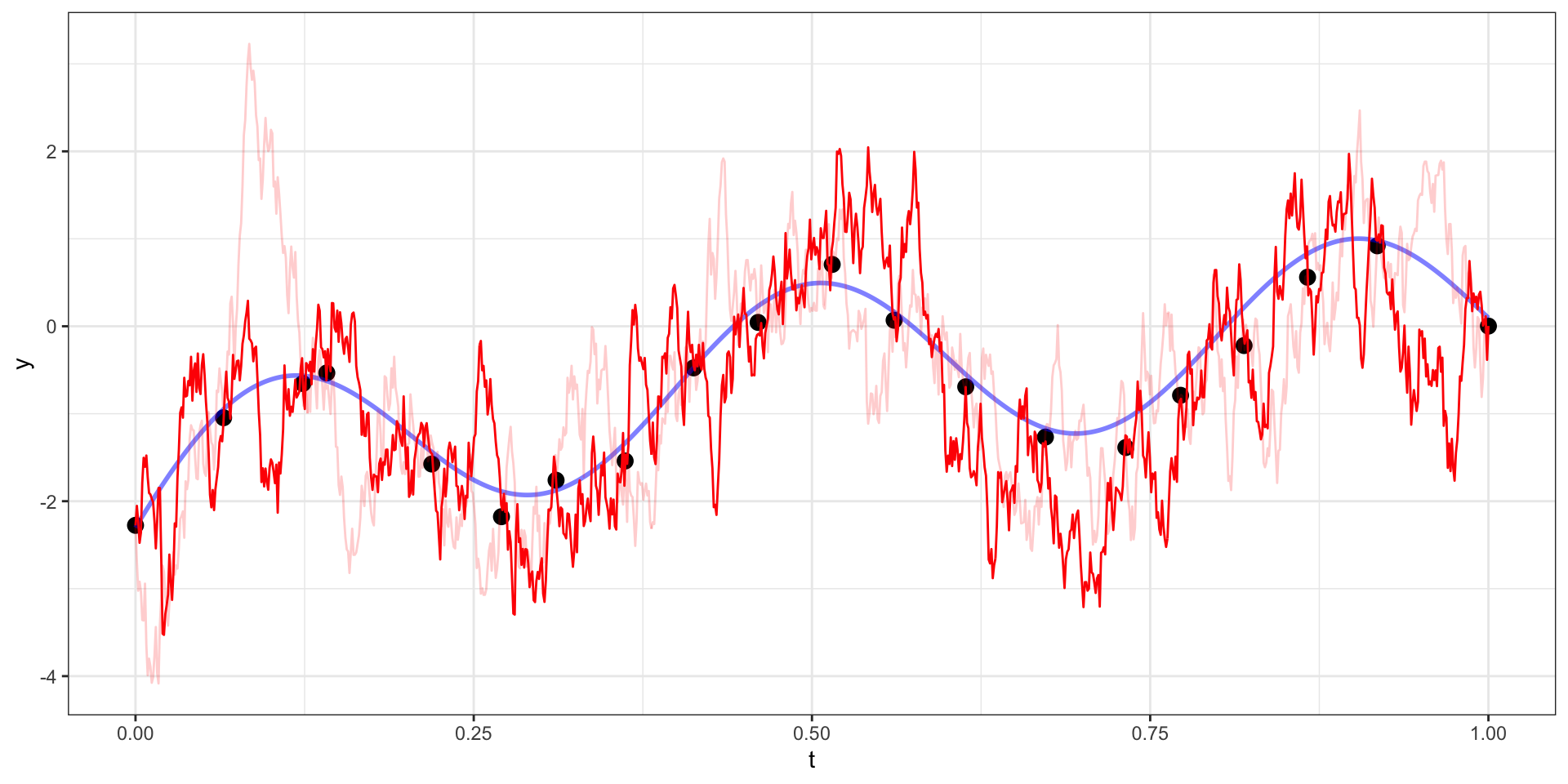
Exponential Covariance
Where \(\sigma=10\), \(l = \sqrt{15}\),
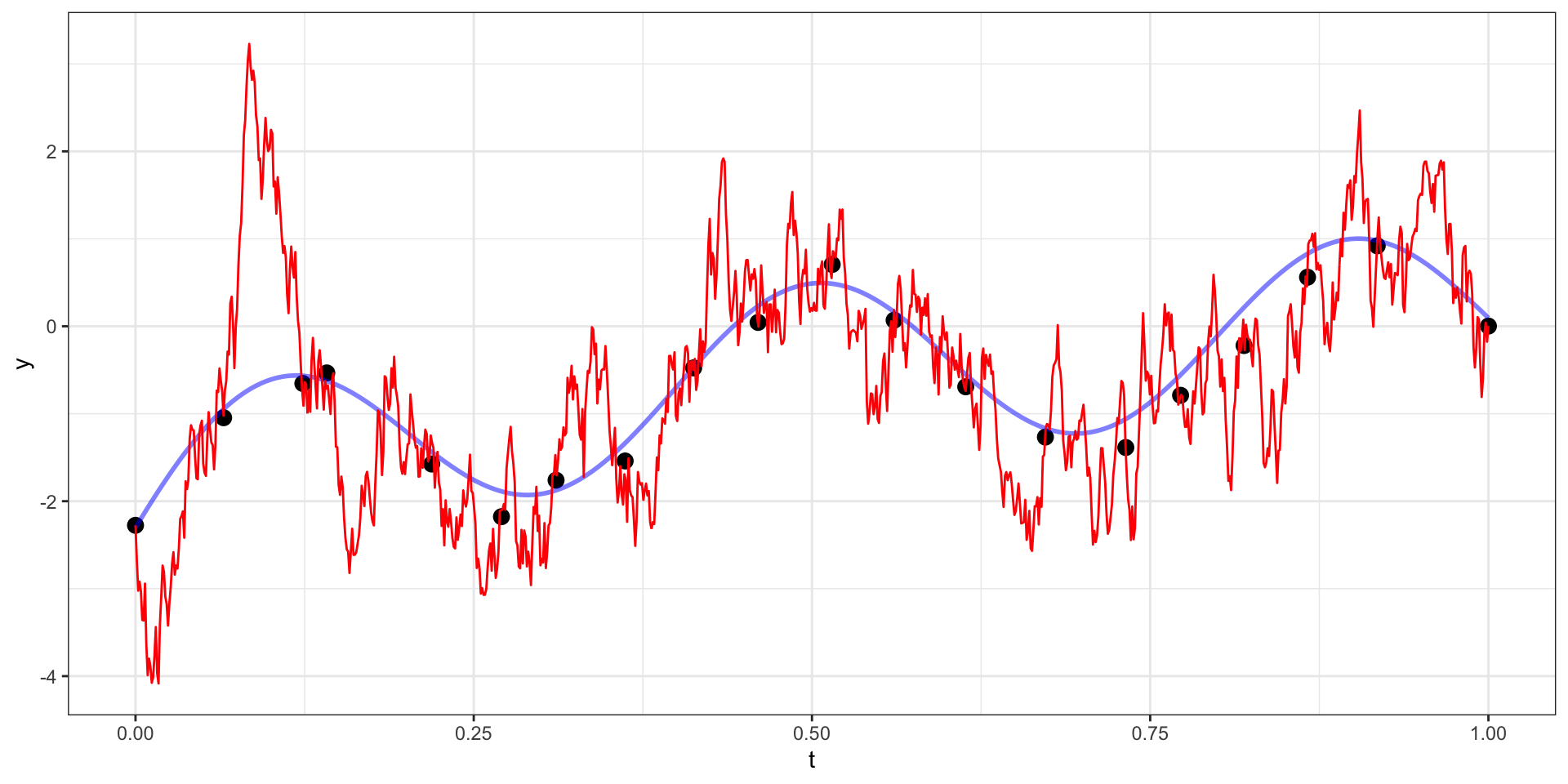
Exponential Covariance - Draw 2
Exponential Covariance - Draw 3
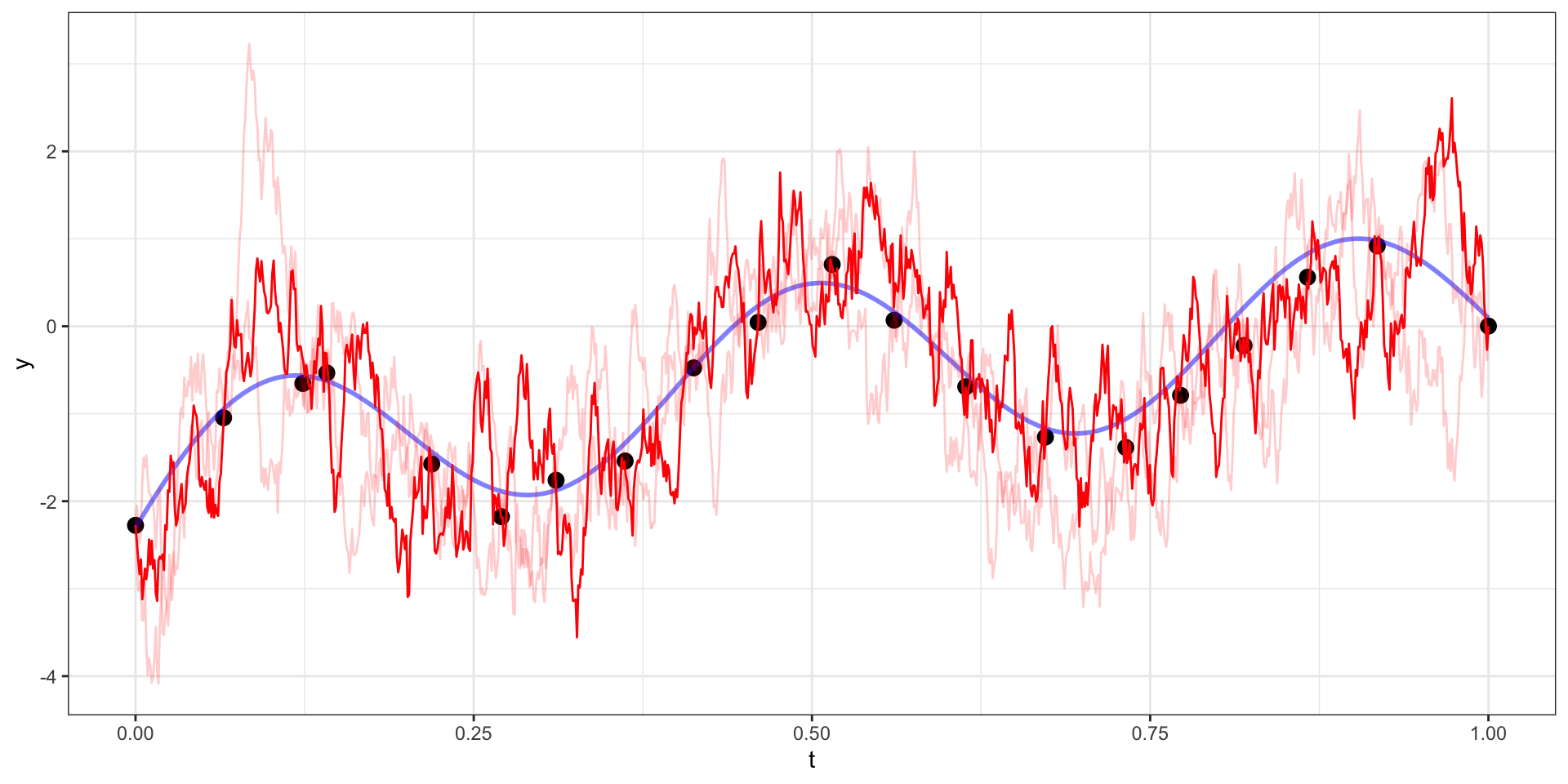
Exponential Covariance - Draw 4
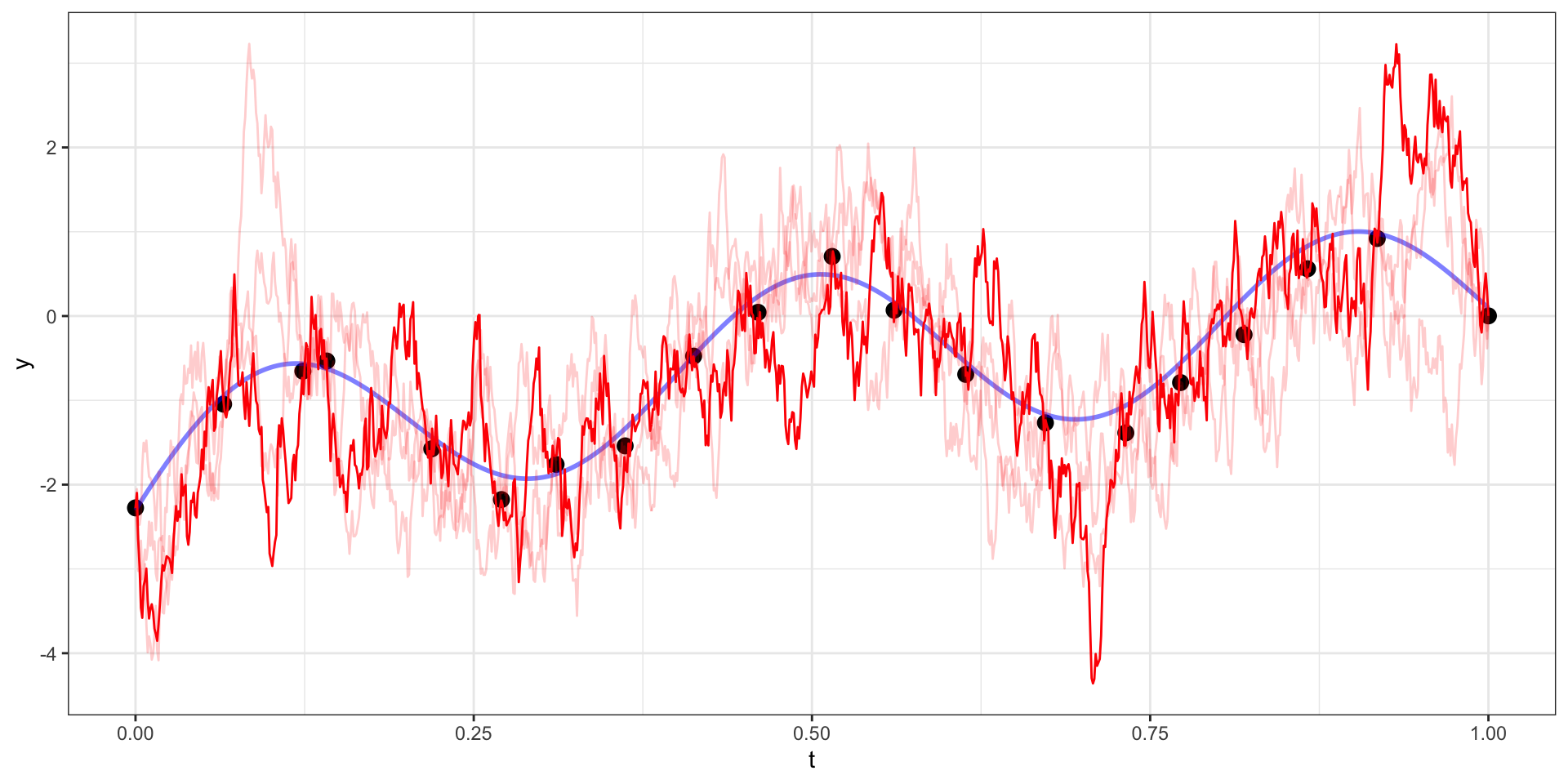
Exponential Covariance - Draw 5
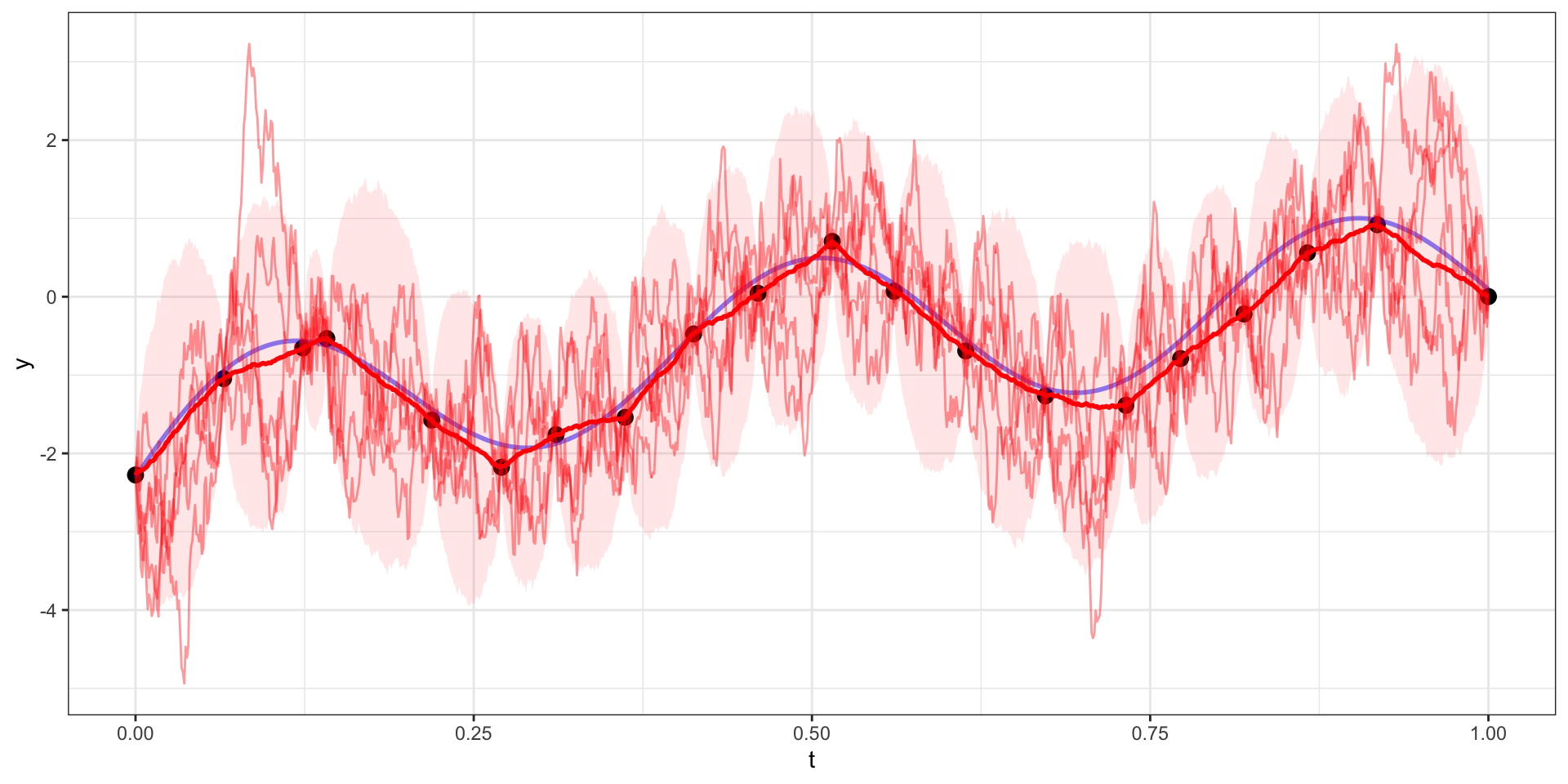
Exponential Covariance - Variability
Powered Exponential Covariance (\(p=1.5\))
Back to the square exponential
Changing the range (\(l\))
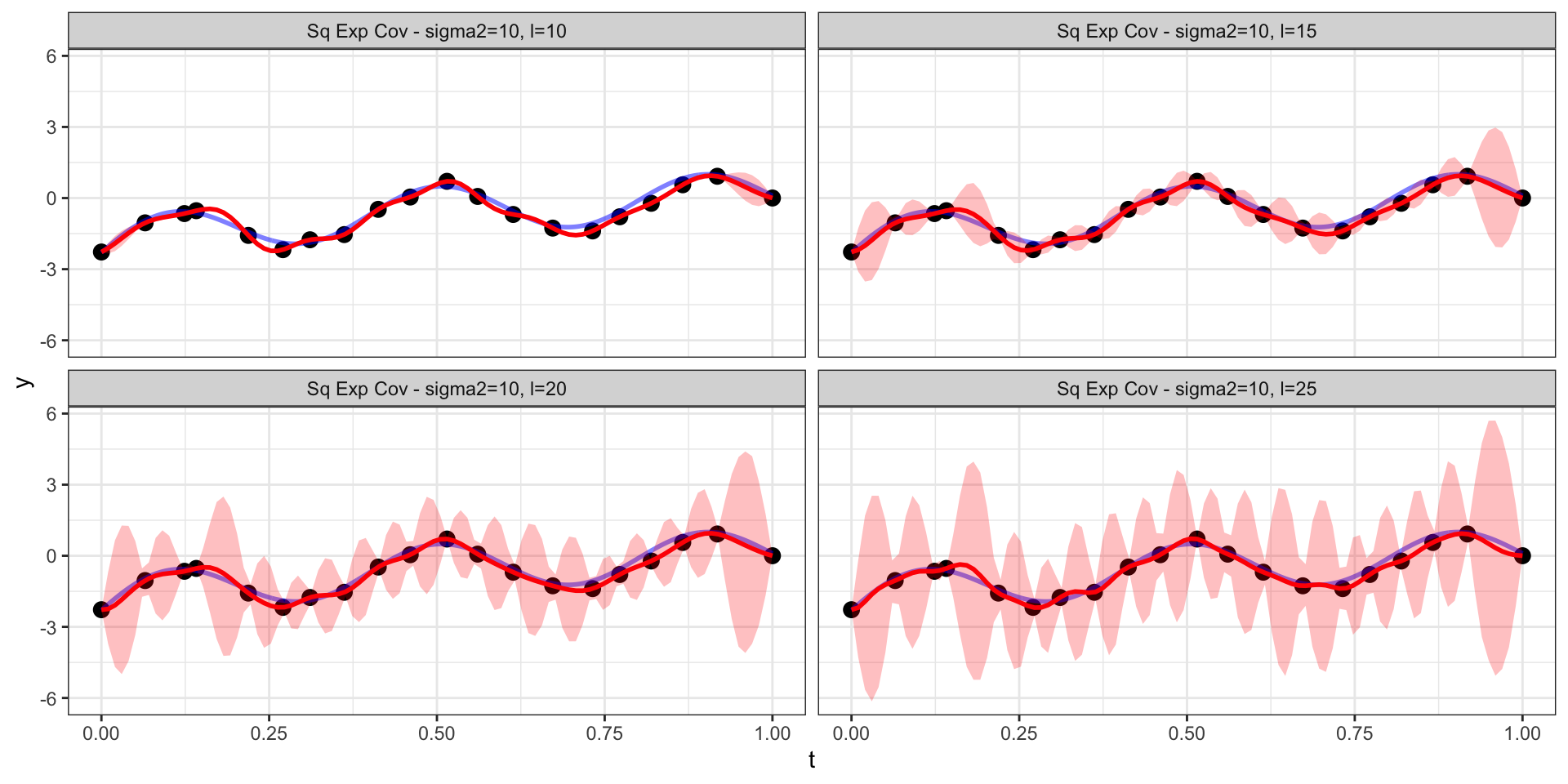
Effective Range
For the square exponential covariance \[ \begin{aligned} Cov(d) &= \sigma^2 \exp\left(-(l \cdot d)^2\right) \\ Corr(d) &= \exp\left(-(l \cdot d)^2\right) \end{aligned} \]
we would like to know, for a given value of \(l\), beyond what distance apart must observations be to have a correlation less than \(0.05\)?
\[ \begin{aligned} \exp\left(-(l \cdot d)^2\right) &< 0.05 \\ -(l \cdot d)^2 &< \log 0.05 \\ l \cdot d &< \sqrt{3} \\ d &< \sqrt{3} / l \end{aligned} \]
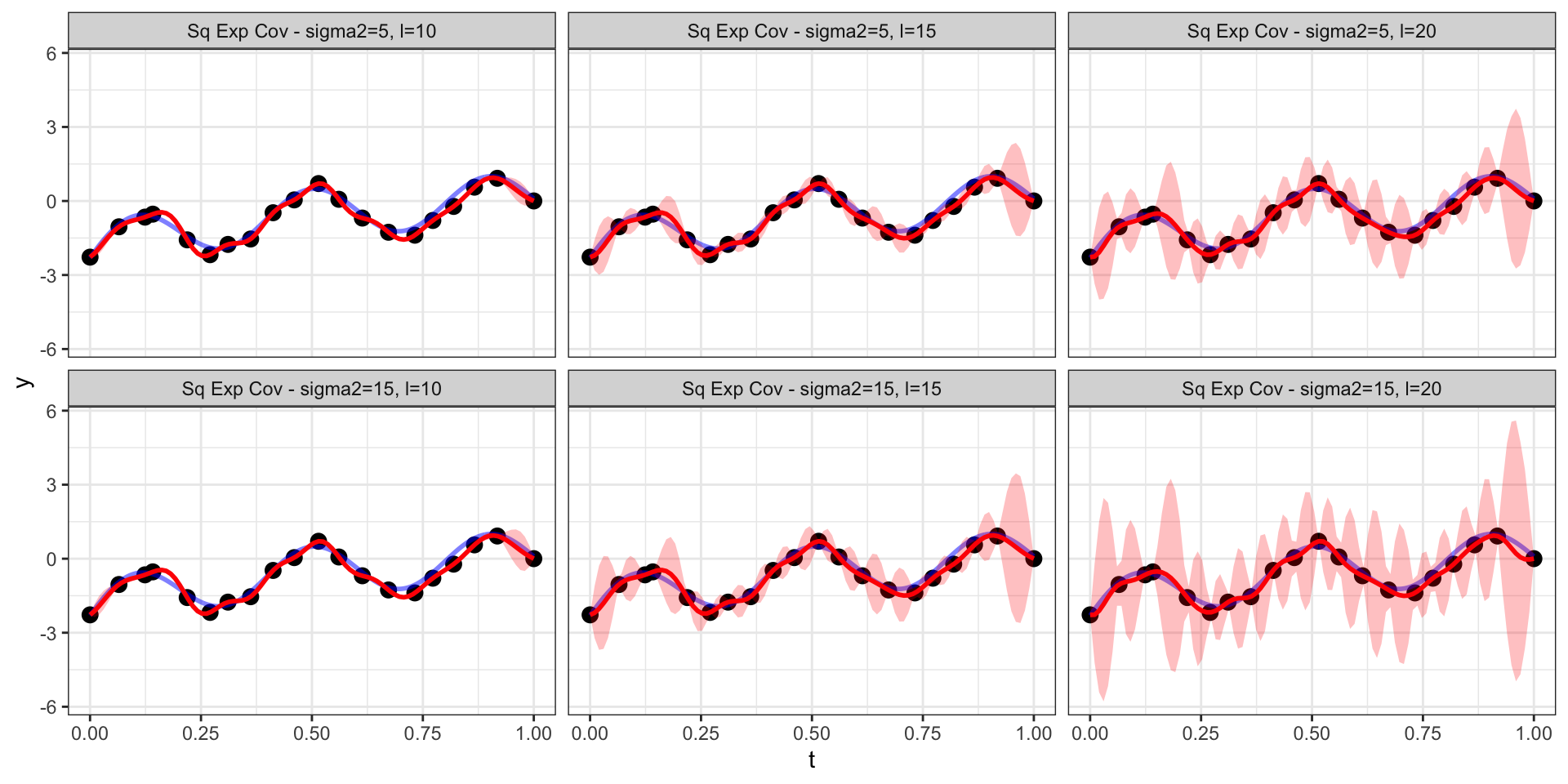
Changing the scale (\(\sigma^2\))
Fitting w/ BRMS
Family: gaussian
Links: mu = identity; sigma = identity
Formula: y ~ gp(t)
Data: d (Number of observations: 20)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
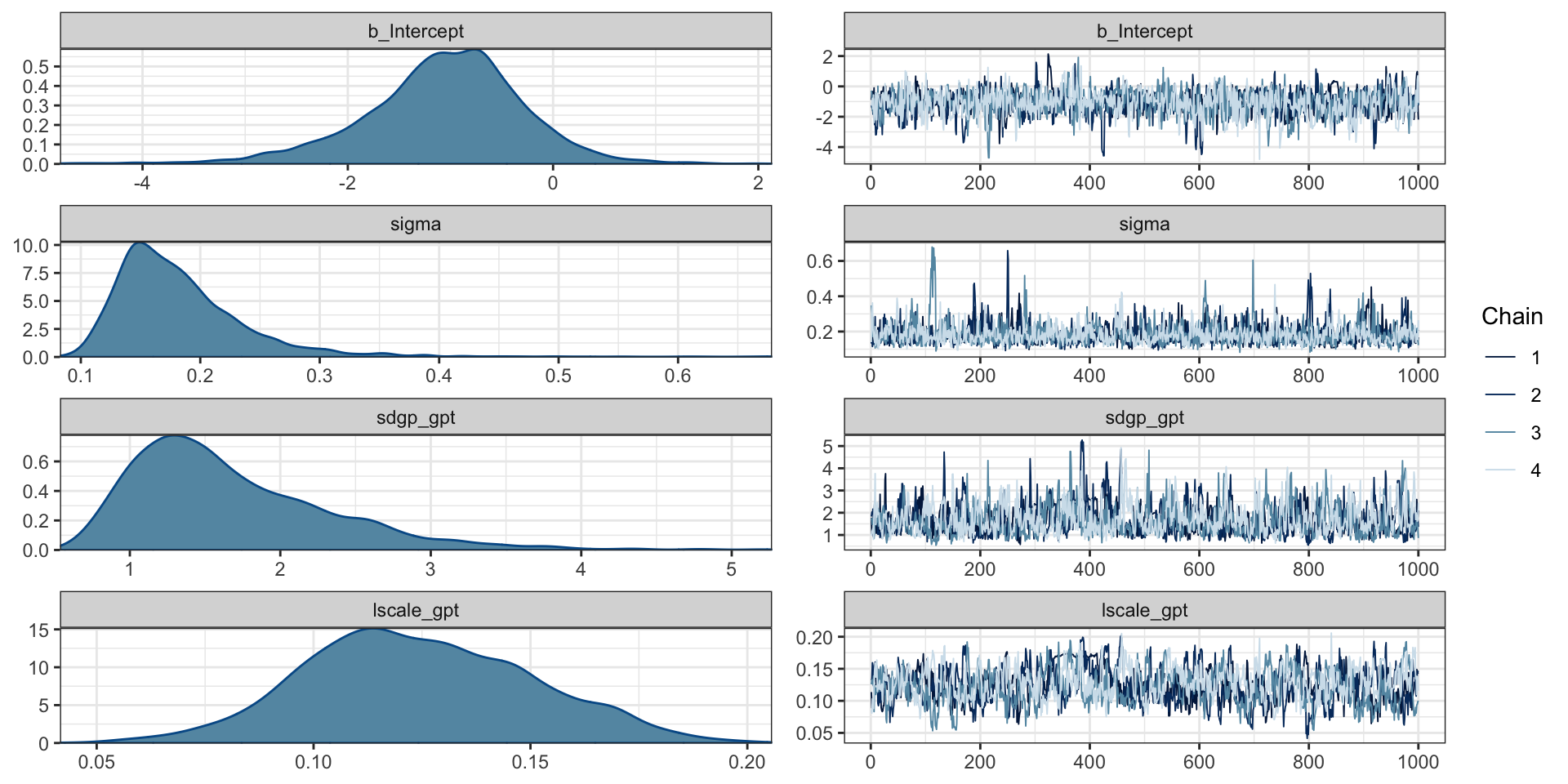
Gaussian Process Terms:
Estimate Est.Error l-95% CI u-95% CI
sdgp(gpt) 1.67 0.67 0.79 3.31
lscale(gpt) 0.12 0.03 0.08 0.18
Rhat Bulk_ESS Tail_ESS
sdgp(gpt) 1.01 390 1098
lscale(gpt) 1.02 262 316
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI
Intercept -1.07 0.79 -2.80 0.36
Rhat Bulk_ESS Tail_ESS
Intercept 1.01 1065 1131
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat
sigma 0.18 0.06 0.11 0.32 1.00
Bulk_ESS Tail_ESS
sigma 914 1180
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Trace plots
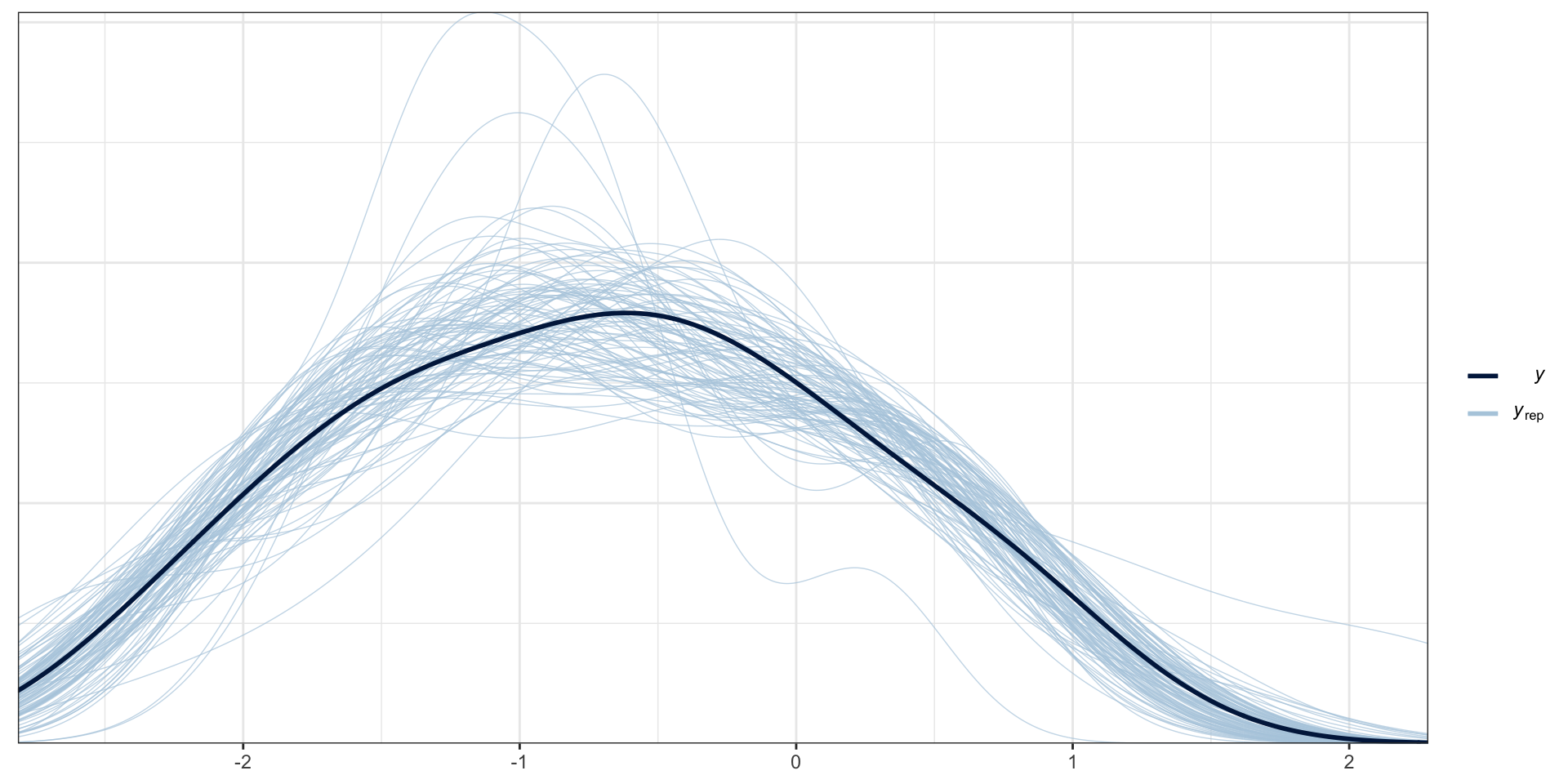
PP Checks
Model predictions
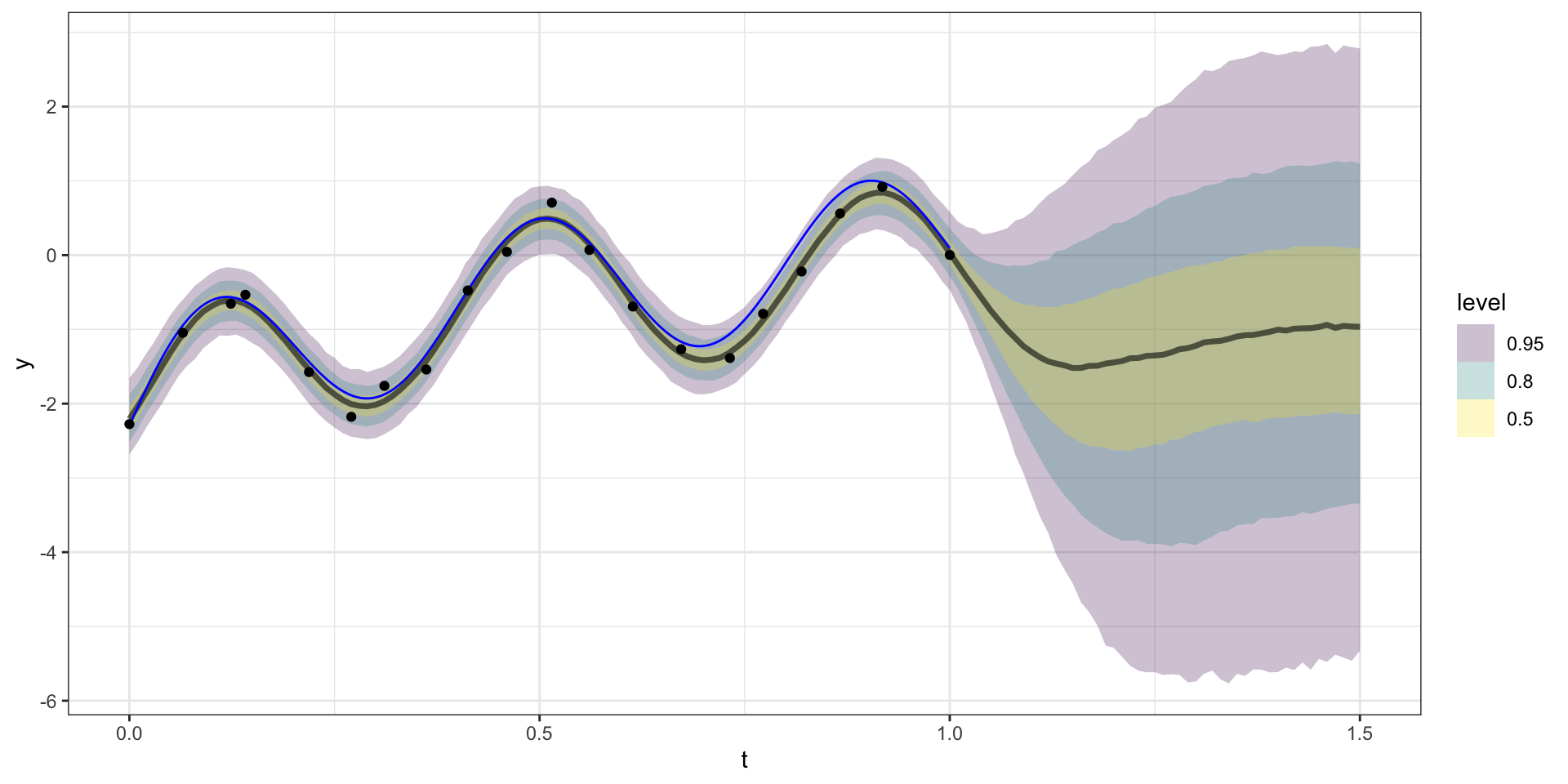
Forecasting
Stan code
// generated with brms 2.18.0
functions {
/* compute a latent Gaussian process
* Args:
* x: array of continuous predictor values
* sdgp: marginal SD parameter
* lscale: length-scale parameter
* zgp: vector of independent standard normal variables
* Returns:
* a vector to be added to the linear predictor
*/
vector gp(data vector[] x, real sdgp, vector lscale, vector zgp) {
int Dls = rows(lscale);
int N = size(x);
matrix[N, N] cov;
if (Dls == 1) {
// one dimensional or isotropic GP
cov = gp_exp_quad_cov(x, sdgp, lscale[1]);
} else {
// multi-dimensional non-isotropic GP
cov = gp_exp_quad_cov(x[, 1], sdgp, lscale[1]);
for (d in 2:Dls) {
cov = cov .* gp_exp_quad_cov(x[, d], 1, lscale[d]);
}
}
for (n in 1:N) {
// deal with numerical non-positive-definiteness
cov[n, n] += 1e-12;
}
return cholesky_decompose(cov) * zgp;
}
/* Spectral density function of a Gaussian process
* with squared exponential covariance kernel
* Args:
* x: array of numeric values of dimension NB x D
* sdgp: marginal SD parameter
* lscale: vector of length-scale parameters
* Returns:
* numeric values of the function evaluated at 'x'
*/
vector spd_cov_exp_quad(data vector[] x, real sdgp, vector lscale) {
int NB = dims(x)[1];
int D = dims(x)[2];
int Dls = rows(lscale);
vector[NB] out;
if (Dls == 1) {
// one dimensional or isotropic GP
real constant = square(sdgp) * (sqrt(2 * pi()) * lscale[1])^D;
real neg_half_lscale2 = -0.5 * square(lscale[1]);
for (m in 1:NB) {
out[m] = constant * exp(neg_half_lscale2 * dot_self(x[m]));
}
} else {
// multi-dimensional non-isotropic GP
real constant = square(sdgp) * sqrt(2 * pi())^D * prod(lscale);
vector[Dls] neg_half_lscale2 = -0.5 * square(lscale);
for (m in 1:NB) {
out[m] = constant * exp(dot_product(neg_half_lscale2, square(x[m])));
}
}
return out;
}
/* how many elements are in a range of integers?
* Args:
* x: an integer array
* start: start of the range (inclusive)
* end: end of the range (inclusive)
* Returns:
* a scalar integer
*/
int size_range(int[] x, int start, int end) {
int out = 0;
for (i in 1:size(x)) {
out += (x[i] >= start && x[i] <= end);
}
return out;
}
/* which elements are in a range of integers?
* Args:
* x: an integer array
* start: start of the range (inclusive)
* end: end of the range (inclusive)
* Returns:
* an integer array
*/
int[] which_range(int[] x, int start, int end) {
int out[size_range(x, start, end)];
int j = 1;
for (i in 1:size(x)) {
if (x[i] >= start && x[i] <= end) {
out[j] = i;
j += 1;
}
}
return out;
}
/* adjust array values to x - start + 1
* Args:
* x: an integer array
* start: start of the range of values in x (inclusive)
* Returns:
* an integer array
*/
int[] start_at_one(int[] x, int start) {
int out[size(x)];
for (i in 1:size(x)) {
out[i] = x[i] - start + 1;
}
return out;
}
}
data {
int<lower=1> N; // total number of observations
vector[N] Y; // response variable
// data related to GPs
int<lower=1> Kgp_1; // number of sub-GPs (equal to 1 unless 'by' was used)
int<lower=1> Dgp_1; // GP dimension
// number of latent GP groups
int<lower=1> Nsubgp_1;
// indices of latent GP groups per observation
int<lower=1> Jgp_1[N];
vector[Dgp_1] Xgp_1[Nsubgp_1]; // covariates of the GP
int prior_only; // should the likelihood be ignored?
}
transformed data {
}
parameters {
real Intercept; // temporary intercept for centered predictors
vector<lower=0>[Kgp_1] sdgp_1; // GP standard deviation parameters
vector<lower=0>[1] lscale_1[Kgp_1]; // GP length-scale parameters
vector[Nsubgp_1] zgp_1; // latent variables of the GP
real<lower=0> sigma; // dispersion parameter
}
transformed parameters {
real lprior = 0; // prior contributions to the log posterior
lprior += student_t_lpdf(Intercept | 3, -0.7, 2.5);
lprior += student_t_lpdf(sdgp_1 | 3, 0, 2.5)
- 1 * student_t_lccdf(0 | 3, 0, 2.5);
lprior += inv_gamma_lpdf(lscale_1[1][1] | 1.82247, 0.11151);
lprior += student_t_lpdf(sigma | 3, 0, 2.5)
- 1 * student_t_lccdf(0 | 3, 0, 2.5);
}
model {
// likelihood including constants
if (!prior_only) {
vector[Nsubgp_1] gp_pred_1 = gp(Xgp_1, sdgp_1[1], lscale_1[1], zgp_1);
// initialize linear predictor term
vector[N] mu = rep_vector(0.0, N);
mu += Intercept + gp_pred_1[Jgp_1];
target += normal_lpdf(Y | mu, sigma);
}
// priors including constants
target += lprior;
target += std_normal_lpdf(zgp_1);
}
generated quantities {
// actual population-level intercept
real b_Intercept = Intercept;
}Sta 344 - Fall 2022