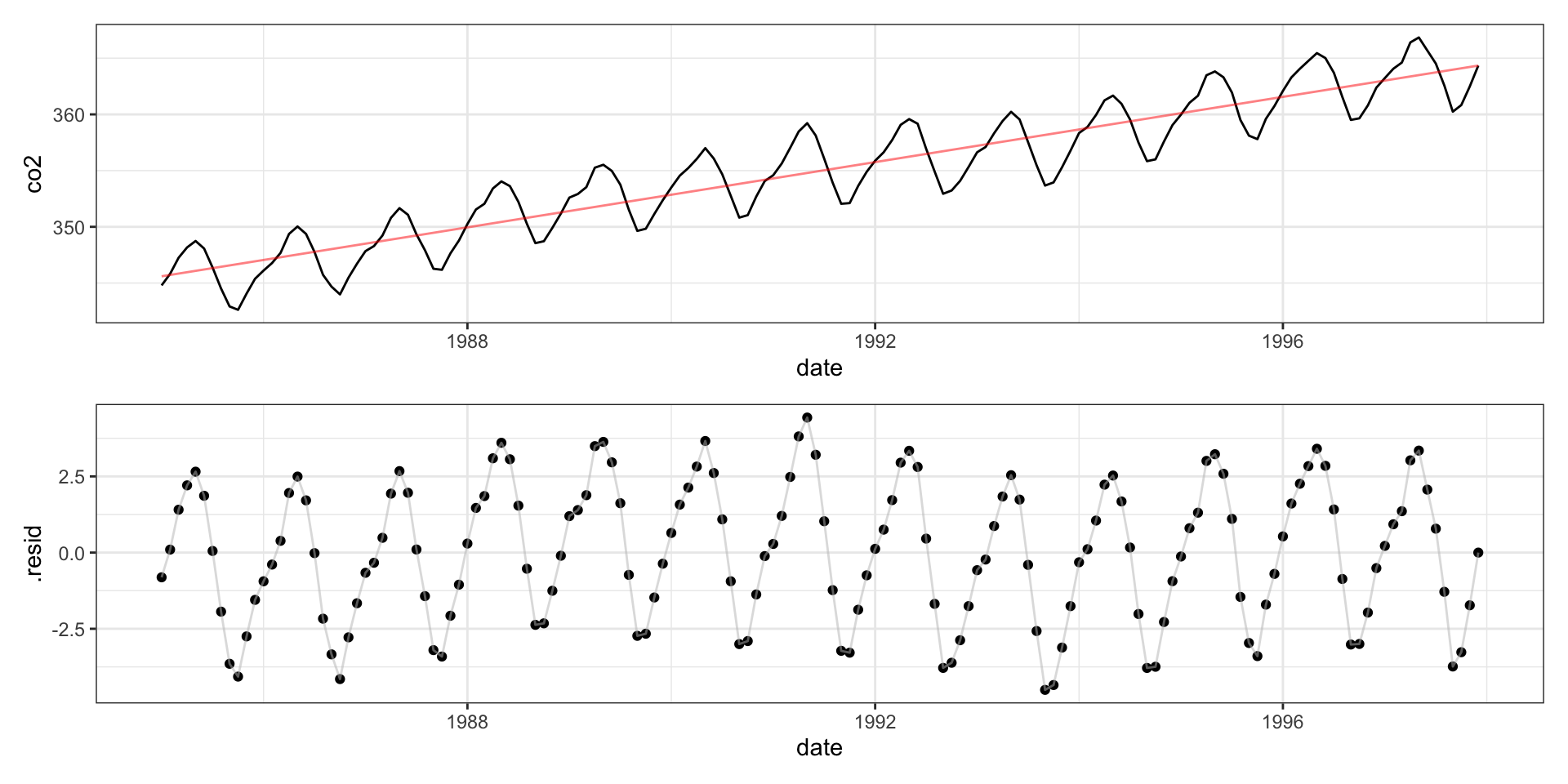
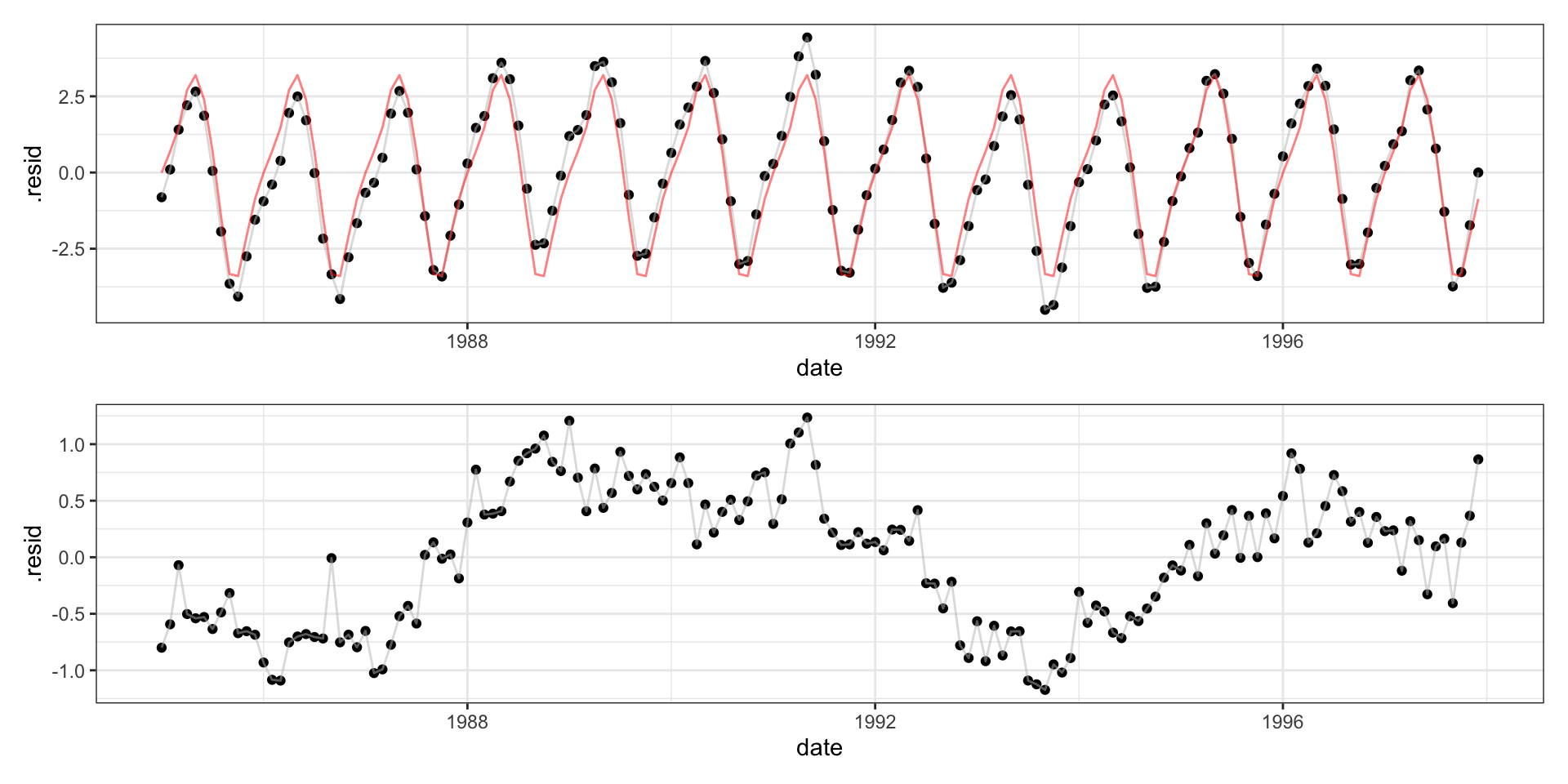
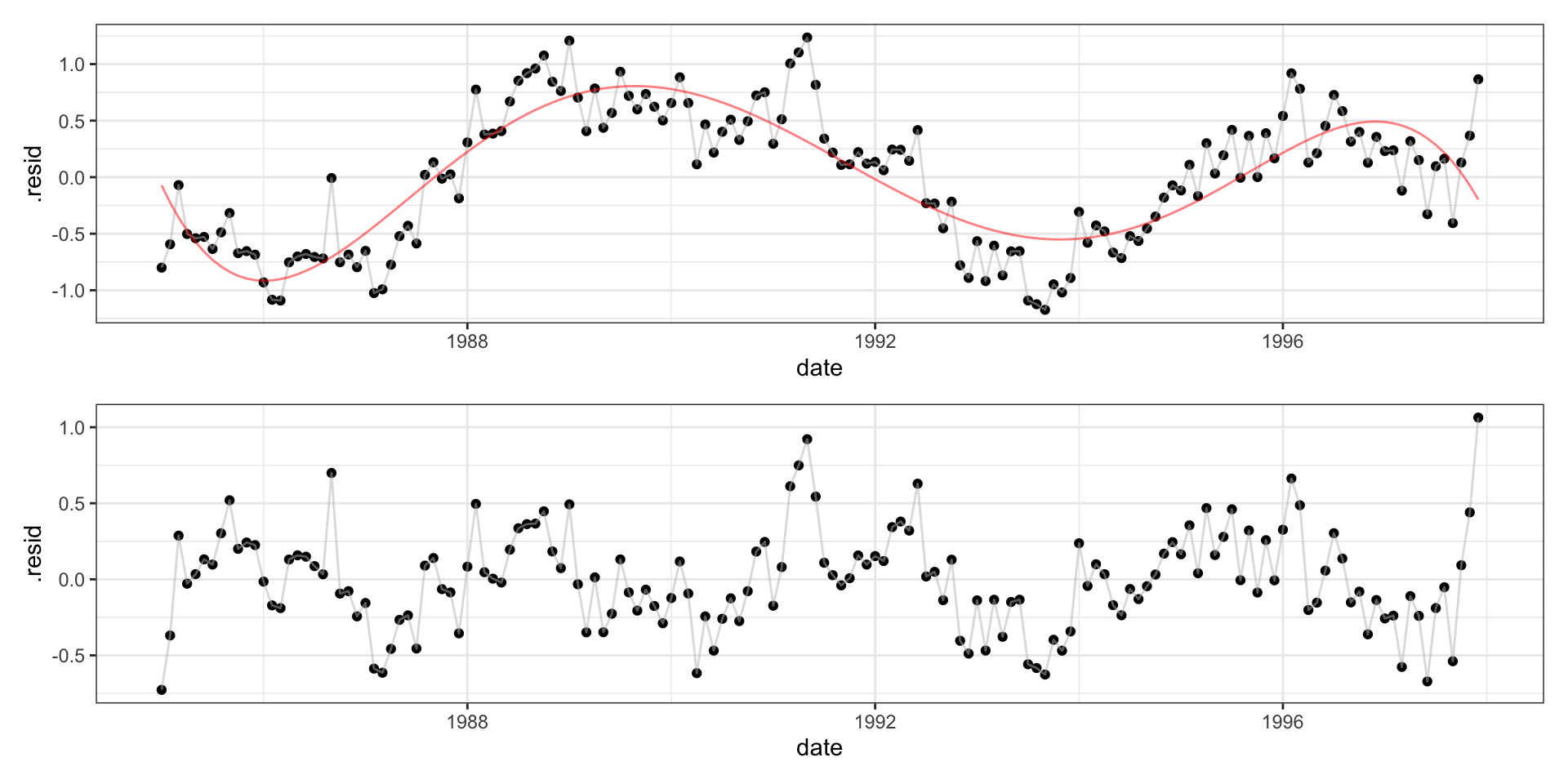
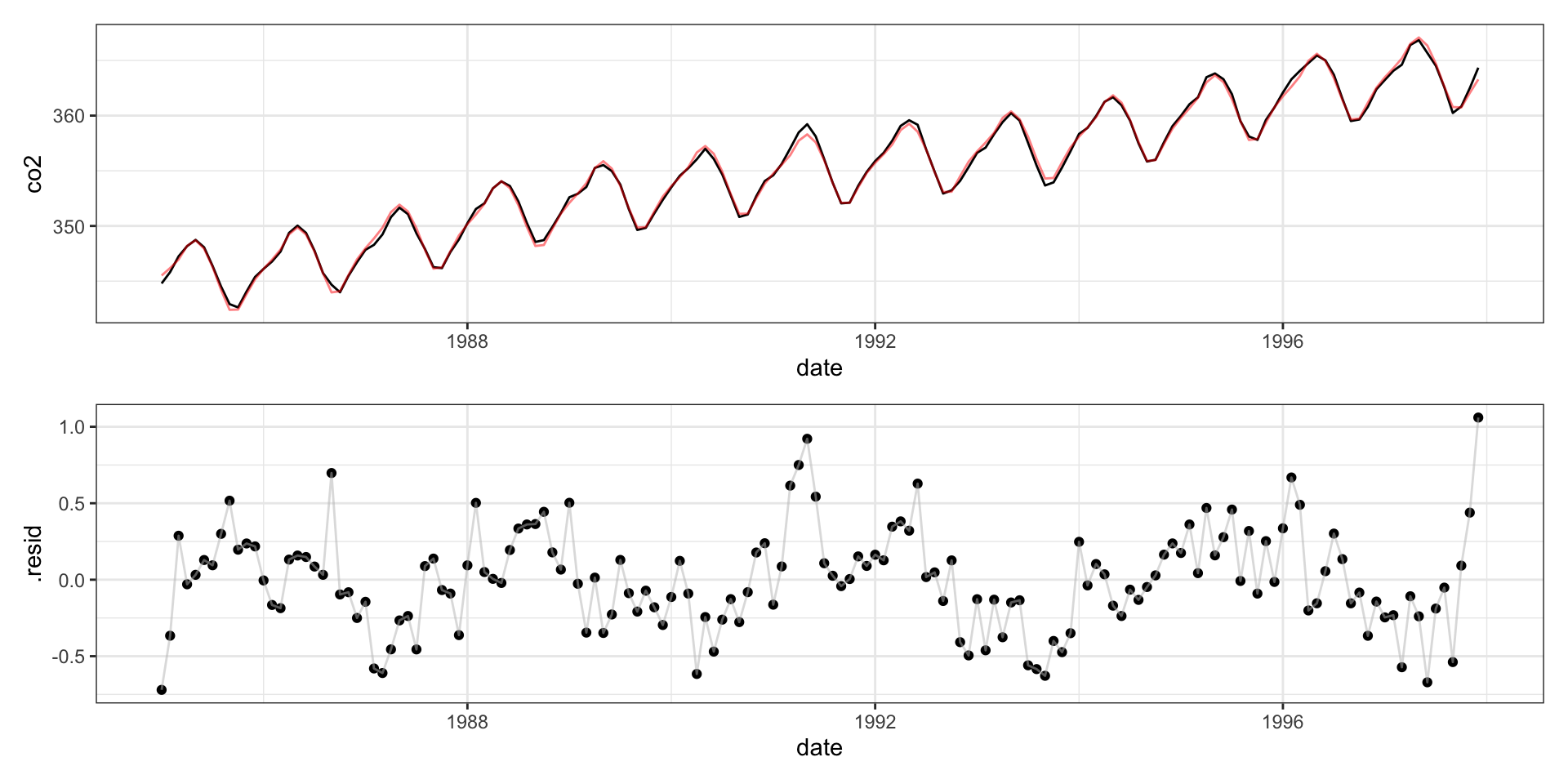
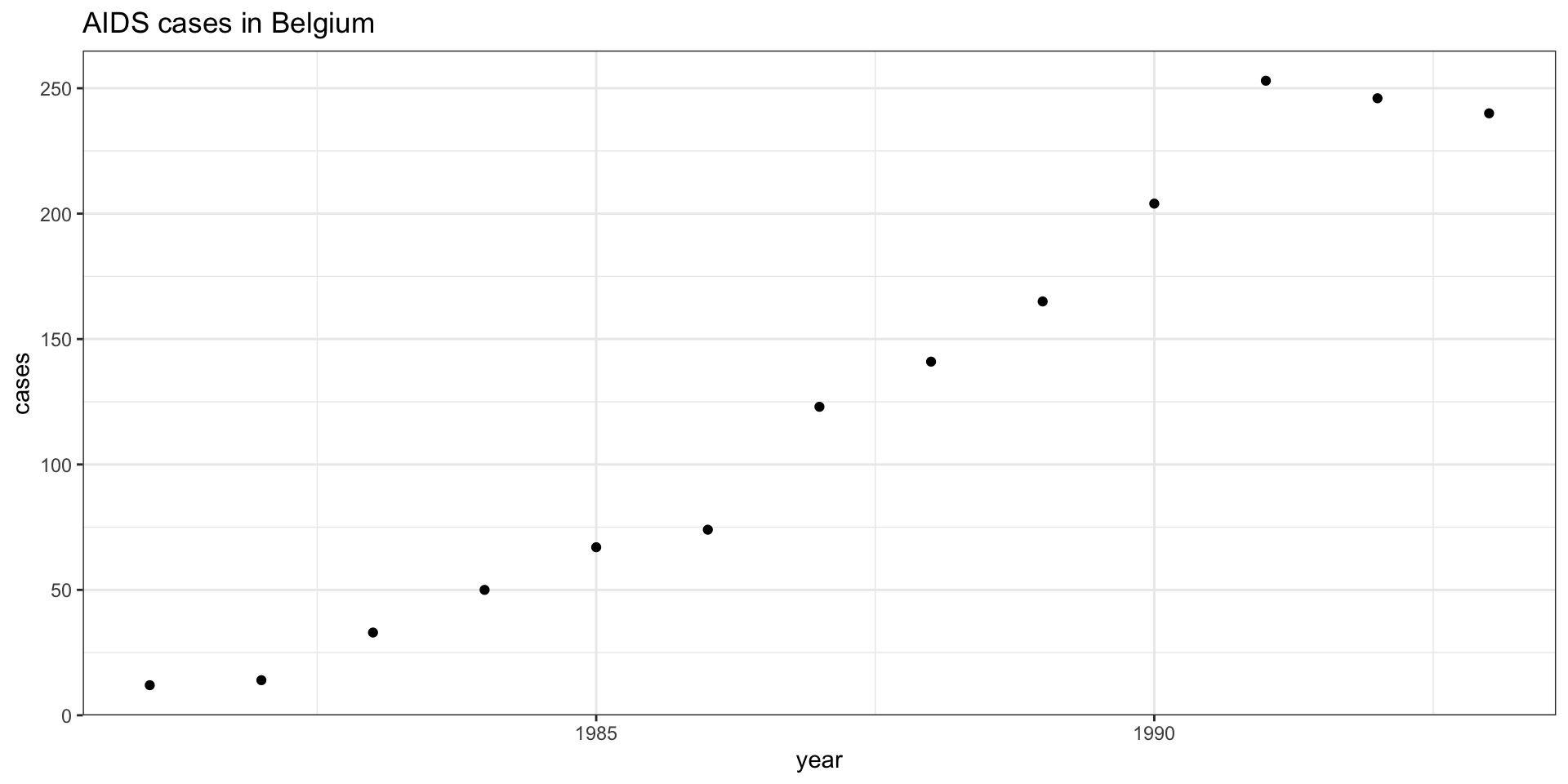
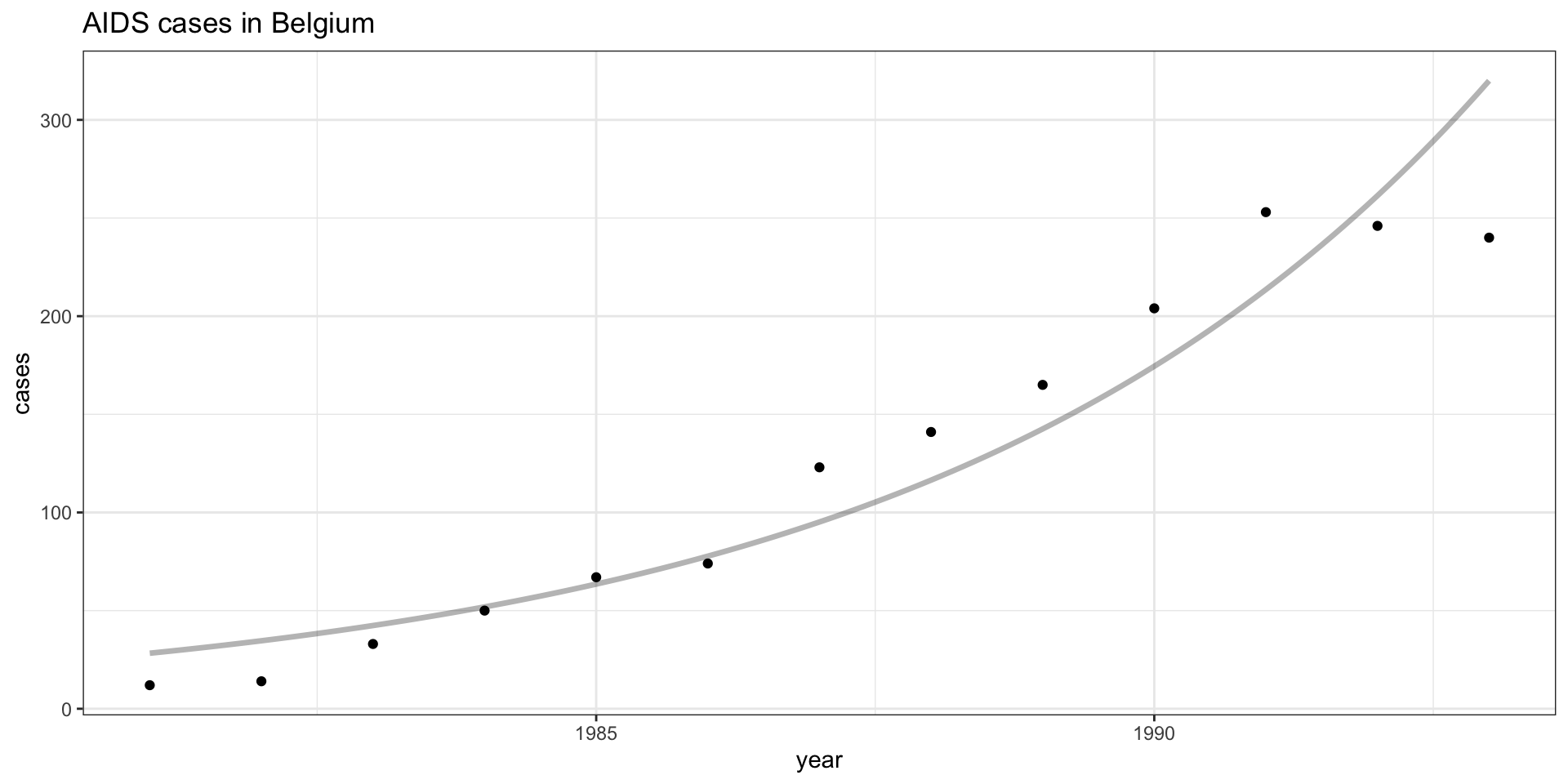
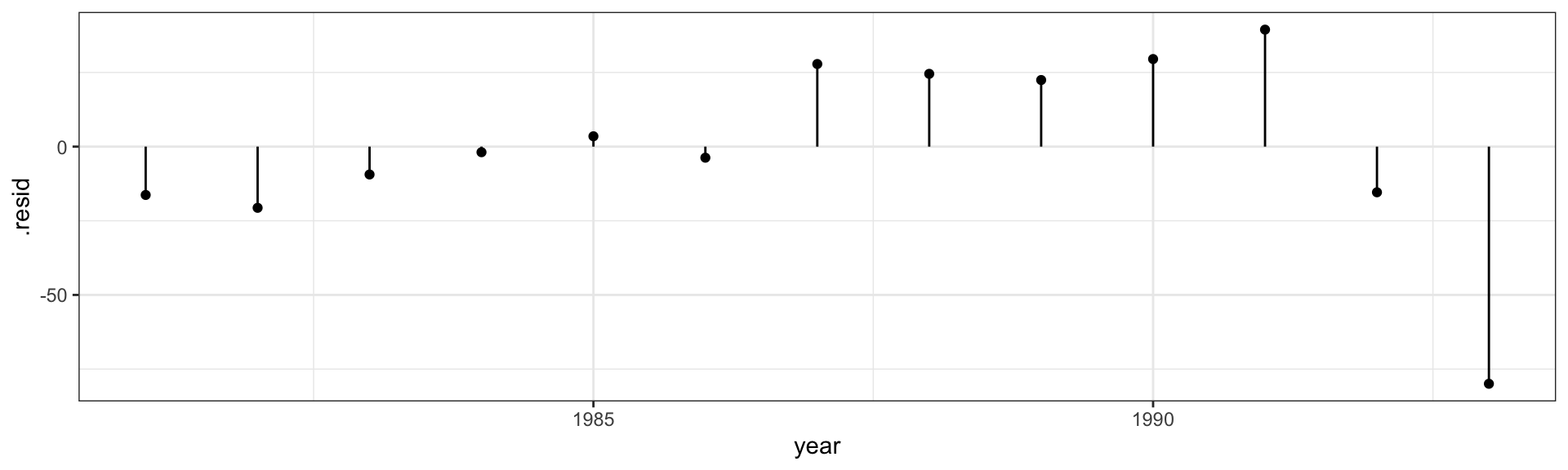
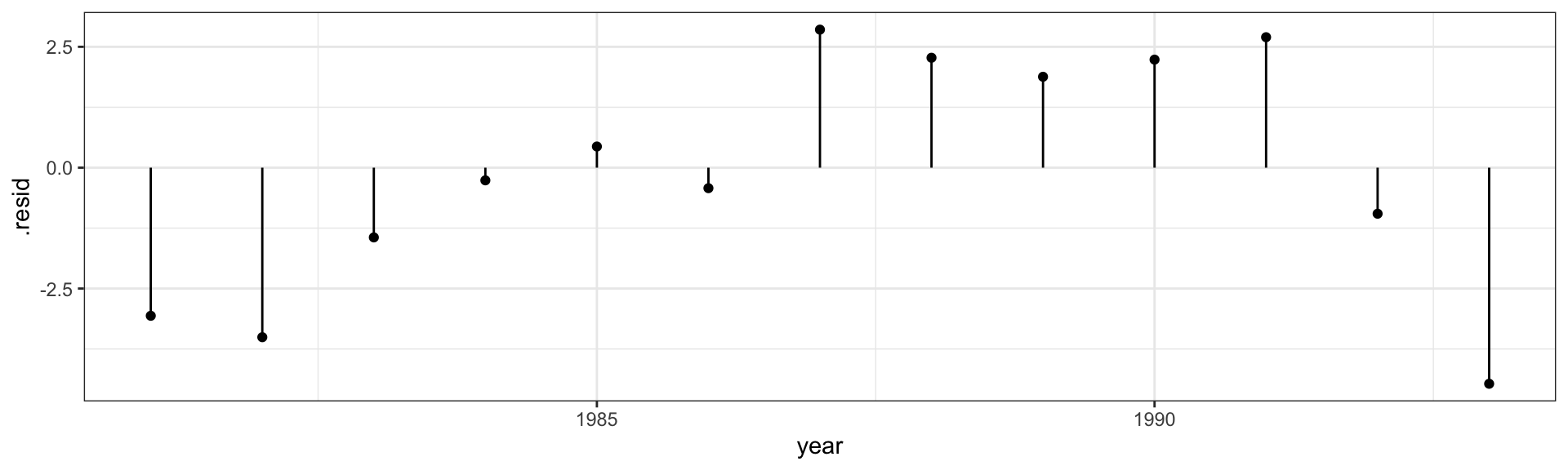
Call:
lm(formula = co2 ~ date + month + poly(date, 5), data = mauna_loa)
Residuals:
Min 1Q Median 3Q Max
-0.72022 -0.19169 -0.00638 0.17565 1.06026
Coefficients: (1 not defined because of singularities)
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.587e+03 1.460e+01 -177.174 < 2e-16 ***
date 1.479e+00 7.334e-03 201.649 < 2e-16 ***
monthAug -4.155e+00 1.346e-01 -30.880 < 2e-16 ***
monthDec -3.566e+00 1.350e-01 -26.404 < 2e-16 ***
monthFeb -2.022e+00 1.345e-01 -15.041 < 2e-16 ***
monthJan -2.729e+00 1.345e-01 -20.286 < 2e-16 ***
monthJul -2.018e+00 1.345e-01 -15.003 < 2e-16 ***
monthJun -3.136e-01 1.345e-01 -2.332 0.021117 *
monthMar -1.233e+00 1.344e-01 -9.175 5.54e-16 ***
monthMay 4.881e-01 1.344e-01 3.631 0.000396 ***
monthNov -4.799e+00 1.349e-01 -35.577 < 2e-16 ***
monthOct -6.102e+00 1.348e-01 -45.282 < 2e-16 ***
monthSep -6.036e+00 1.346e-01 -44.832 < 2e-16 ***
poly(date, 5)1 NA NA NA NA
poly(date, 5)2 -1.920e+00 3.427e-01 -5.602 1.09e-07 ***
poly(date, 5)3 3.920e+00 3.451e-01 11.358 < 2e-16 ***
poly(date, 5)4 8.946e-01 3.428e-01 2.609 0.010062 *
poly(date, 5)5 -4.340e+00 3.462e-01 -12.535 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.3427 on 139 degrees of freedom
Multiple R-squared: 0.997, Adjusted R-squared: 0.9966
F-statistic: 2872 on 16 and 139 DF, p-value: < 2.2e-16